 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_05738-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 720aa MW: 83372.7 Da PI: 7.869 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 57.8 | 3.7e-18 | 62 | 147 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
+fY++YA++ GF v ++++++ +++ t t+ C+++gk++ + k+ + +++++tr++C ak + +++ +++++t++ l+HnH+++p
TRIUR3_05738-P1 62 RFYKAYARKKGFGVIRRTTRHG-EDKMLTYFTLACNRQGKAQYSAKN---SFKPNPSTRMQCPAKSNFSRR-GENFCITTVTLDHNHPISP 147
5**************9998887.7888****************9999...999*****************9.9**************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.5E-15 | 62 | 147 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.8E-34 | 267 | 361 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.433 | 556 | 594 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 4.4E-4 | 564 | 590 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 2.9E-4 | 569 | 596 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 720 aa Download sequence Send to blast |
MEPDLTLDEV AEQNRSAPDE EDEDAFVVEE IGDSSDDRQA EPTVALEPKK GMMFSSEDDA 60 VRFYKAYARK KGFGVIRRTT RHGEDKMLTY FTLACNRQGK AQYSAKNSFK PNPSTRMQCP 120 AKSNFSRRGE NFCITTVTLD HNHPISPSKA RFLRCHKKVD LHSKRRLELN DQAGARMNKN 180 FGSPVMEAGW YGNLEFGEKE CTNYLQEKRS LKLGAGDAHA LYRYFLHMQS KDPAFFHVMD 240 VAEDGRLRNV FWADARSRAA YESYWDVITF DTTYLTNKYT IPLATFVGIN HHGESVLLGC 300 GLLSNEDTET FVWLFKSWLC CMSCKPPNAI ITDQCKPMQN AIEEVFPQAR HRWCACHIMK 360 KIPENLNGYE NIKSTLSNVV YDSLTKHGFD KAWVEMINKY DLQENEWLAG LYDNKNRWVP 420 AYVKDTFWAG MSSTKRSESV NAFFDGYVNA RTTLKQFVEQ YENLLRDKVE KENKADSKSF 480 QQQIPCITHY DFERQFQAAY TTAKFKEFQD QLRGKIYCYP TQLNKEGSIF TFGVREDSKI 540 FCEGEDGKGK EKRVILEFIV LFNQGECDVQ CMCRLFEFRG ILCSHIISVL ALMEITDVPS 600 RYILQRWRKD IKRKHTFIRC SYDDMIDTQV VQRYDNLCKR SHELAENGAE SDALHDLVMD 660 GLNELQSKID AYCARYNEKE KKEKIVLSPI PIRSVGCPPS KRKESKVDQV VKKLRAKKQQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
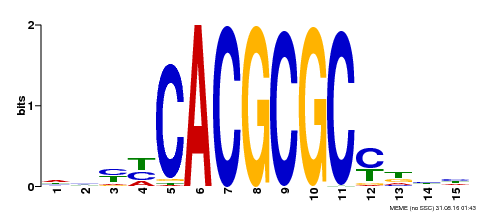
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ389711 | 0.0 | HQ389711.1 Triticum aestivum clone UCDTA00162 genomic sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020171727.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| Refseq | XP_020171728.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| Refseq | XP_020171729.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| Refseq | XP_020171730.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| Refseq | XP_020171731.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1-like isoform X1 | ||||
| TrEMBL | T1LLP8 | 0.0 | T1LLP8_TRIUA; Uncharacterized protein | ||||
| STRING | TRIUR3_05738-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP18 | 32 | 967 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 1e-131 | FAR1 family protein | ||||



