 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_10045-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 906aa MW: 101875 Da PI: 7.8073 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.9 | 8.7e-06 | 557 | 579 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cg +F+ + +Lk+H+++H
TRIUR3_10045-P1 557 HTCEECGACFRKPAHLKQHMQSH 579
79*******************99 PP
| |||||||
| 2 | zf-C2H2 | 20.6 | 1.2e-06 | 625 | 649 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ s+ rk++L+rH+ tH
TRIUR3_10045-P1 625 FACPleDCPFSYIRKDHLNRHMLTH 649
78**********************9 PP
| |||||||
| 3 | zf-C2H2 | 15.7 | 4.4e-05 | 654 | 679 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp Cgk+Fs k n +rH + H
TRIUR3_10045-P1 654 FTCPleGCGKRFSIKANIQRHVKEmH 679
89********************9988 PP
| |||||||
| 4 | zf-C2H2 | 15.3 | 5.8e-05 | 698 | 720 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
C+ C+k F+ +s+Lk+H +H
TRIUR3_10045-P1 698 CQeeGCKKAFKYPSQLKKHEESH 720
77779**************9988 PP
| |||||||
| 5 | zf-C2H2 | 15.2 | 6.4e-05 | 787 | 811 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C sFs+ksnL++H++ H
TRIUR3_10045-P1 787 KCSfdGCECSFSTKSNLNKHMKAcH 811
799999****************955 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51375 | 5.481 | 35 | 65 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.879 | 66 | 100 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 3.2E-8 | 67 | 167 | IPR011990 | Tetratricopeptide-like helical domain |
| Pfam | PF01535 | 0.6 | 68 | 97 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.353 | 136 | 166 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 2.6E-4 | 141 | 164 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 5.4E-4 | 141 | 163 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.627 | 167 | 197 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.0043 | 169 | 195 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.986 | 198 | 232 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 4.3E-4 | 200 | 229 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 2.2E-4 | 200 | 233 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.079 | 268 | 298 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 3.2E-8 | 269 | 299 | IPR011990 | Tetratricopeptide-like helical domain |
| Pfam | PF01535 | 0.0039 | 273 | 296 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF13041 | 7.4E-11 | 298 | 346 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 12.233 | 299 | 333 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 2.3E-8 | 301 | 335 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 8.824 | 334 | 364 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 6.138 | 370 | 400 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.42 | 373 | 397 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 3.2E-8 | 400 | 464 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 5.415 | 402 | 432 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 6.95 | 436 | 470 | IPR002885 | Pentatricopeptide repeat |
| SMART | SM00355 | 42 | 511 | 534 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.3 | 557 | 584 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-5 | 557 | 582 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0053 | 557 | 579 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 559 | 579 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-6 | 623 | 651 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.515 | 625 | 654 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 7.4E-4 | 625 | 649 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 627 | 649 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.0E-9 | 635 | 686 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.3E-6 | 652 | 676 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 12.217 | 654 | 684 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.0E-4 | 654 | 679 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 656 | 679 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.245 | 696 | 721 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.039 | 696 | 720 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.0E-6 | 698 | 720 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 698 | 720 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5 | 728 | 753 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 730 | 753 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 32 | 756 | 777 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.24E-8 | 771 | 828 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.6E-6 | 771 | 808 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.198 | 786 | 816 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0016 | 786 | 811 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 788 | 811 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.2 | 817 | 843 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.642 | 817 | 843 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 819 | 843 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 906 aa Download sequence Send to blast |
MNRSVCHHLL AQCRSLRELE RIHARAVTHG LHPCNQSISC KLFRCYADFG RPAEARRLFG 60 EIPCPDLVSF TSLMSLHLQL NQHHEAMSAF SRAIASGHRP DGFVVVGALS ASGGAGDLRA 120 GLGIHCLIFR CGLGSELVVG NALVDMYSRC GRFEGALGVF DEMAVKDEVT WGSMLHGYMK 180 CVGVDSALSF FDRMPVKSTV SWTALVTGHV QAKQPIRALE IFGRMVLEGH RPTHVTIVGV 240 LSACADIGSL DLGRVIHGYG SKFNISTNII VSNALMDMYA KSGSIEMAFA VFEEVLVKDA 300 FTWTTMISSF TVQGDGIKAL ELFEDMLRSG IVPNSVTFVS VLSACSHAGL IQQGRELFDK 360 MRRIYNIKPQ LEHYGCIVDL LGRGGLLEEA EALIYDMDVE PDTVIWRSLL SACLVHGNDR 420 LAEIAGREII KREPGDDGVY VLLWNIYASS DRWKQALDIR KQMLTRKIFK KPGCSWIEVD 480 GGVHEFLMSS GDGIDGDEKS EETHGKDIRR IKCEFCTVVR SKMYLIRAHM VAQHKDELDA 540 SEIYDSNGEK VVYGVGHTCE ECGACFRKPA HLKQHMQSHS KEVSLRRVRG SNNKMSSQKA 600 STILHWFSAR YGDNTRKNKL IGRSFACPLE DCPFSYIRKD HLNRHMLTHE GKLFTCPLEG 660 CGKRFSIKAN IQRHVKEMHE DEHECKIDDT KSNQQVICQE EGCKKAFKYP SQLKKHEESH 720 AKLDYVEVVC CEPGCLKTFT NAKCLRAHNQ SCHLYTHCDI CGEKHLRKNI KRHLRAHDEV 780 HSTERLKCSF DGCECSFSTK SNLNKHMKAC HDQVRPFECR VAGCGKTFPY KHVRDNHEKS 840 SCHVYVEGDY EEMDEQLRSR RGGGRKRSAV TVETLTRKRV TIPSEVSSLD DGAEYMRWML 900 SDGDDS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5iww_D | 9e-54 | 9 | 412 | 4 | 330 | PLS9-PPR |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
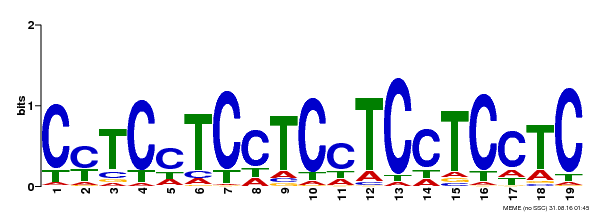
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK374527 | 0.0 | AK374527.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3068C12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020181266.1 | 0.0 | putative pentatricopeptide repeat-containing protein At5g59200, chloroplastic | ||||
| TrEMBL | T1LY66 | 0.0 | T1LY66_TRIUA; Uncharacterized protein | ||||
| STRING | TRIUR3_10045-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-106 | transcription factor IIIA | ||||



