 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_11780-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 289aa MW: 31545.3 Da PI: 5.094 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.7 | 2.3e-19 | 27 | 79 | 4 | 56 |
-SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 4 RttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+ +f++eq++ Le++F+ + ++ +++ +LA++lgL+ rqV +WFqN+Ra++k
TRIUR3_11780-P1 27 KKRFSEEQIKSLESMFATQTKLEPRQKLQLARELGLQPRQVAIWFQNKRARWK 79
456*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 122.5 | 2e-39 | 26 | 115 | 2 | 91 |
HD-ZIP_I/II 2 kkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
+k+r+s+eq+k+LE++F +++kLep++K +lareLglqprqva+WFqn+RAR+k+kqlE++y+aL++ ydal ++ ++L+k++++L +++
TRIUR3_11780-P1 26 RKKRFSEEQIKSLESMFATQTKLEPRQKLQLARELGLQPRQVAIWFQNKRARWKSKQLERQYAALRDDYDALLSSYDQLKKDKQALLNQV 115
8*************************************************************************************9886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-19 | 17 | 81 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 16.876 | 21 | 81 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.31E-18 | 21 | 83 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 3.6E-16 | 24 | 85 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.08E-17 | 26 | 82 | No hit | No description |
| Pfam | PF00046 | 1.0E-16 | 27 | 79 | IPR001356 | Homeobox domain |
| PRINTS | PR00031 | 1.0E-5 | 52 | 61 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 56 | 79 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.0E-5 | 61 | 77 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.2E-11 | 81 | 115 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MEQGEEDGDW MVEPAAGKKG GAMIDRKKRF SEEQIKSLES MFATQTKLEP RQKLQLAREL 60 GLQPRQVAIW FQNKRARWKS KQLERQYAAL RDDYDALLSS YDQLKKDKQA LLNQVCLYST 120 TYQVLVSPCS PAGGGVRRVL VVPVANARST GSVQLEKLAE MLREPGGAKC GDNAGAAARD 180 DVRLAVAGMS MKDEFVDAGG ASKLYSASEG CGGSGKLSLF GEEDDDAGLF LRPSLQLPTA 240 HDGGFTASGP AEYQQQSPSS FPFHSSWPSS AAEQTCSSSQ WWEFESPSE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:10732669}. | |||||
| UniProt | Probable transcription factor that binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:10732669}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00650 | PBM | Transfer from LOC_Os09g35910 | Download |
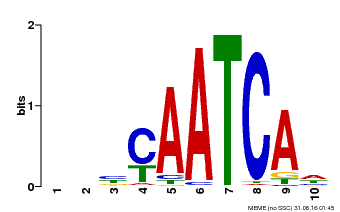
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT224374 | 0.0 | KT224374.1 Triticum aestivum cultivar RAC875 HDZipI-3 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020148448.1 | 1e-162 | homeobox-leucine zipper protein HOX6-like | ||||
| Swissprot | Q651Z5 | 7e-86 | HOX6_ORYSJ; Homeobox-leucine zipper protein HOX6 | ||||
| Swissprot | Q9XH35 | 7e-86 | HOX6_ORYSI; Homeobox-leucine zipper protein HOX6 | ||||
| TrEMBL | M7YI40 | 0.0 | M7YI40_TRIUA; Homeobox-leucine zipper protein HOX6 | ||||
| STRING | TRIUR3_11780-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7873 | 34 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46680.1 | 5e-40 | homeobox 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



