 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_12123-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 600aa MW: 62860.2 Da PI: 7.1062 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 35.1 | 3.3e-11 | 261 | 317 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + ++ k + g ++ +e+Aa+a++ a++k++g
TRIUR3_12123-P1 261 SIYRGVTRHRWTGRYEAHLWDnSCR-RegQTrKGRQ-GGYDKEEKAARAYDLAALKYWG 317
57*******************4444.2455535555.779999**************98 PP
| |||||||
| 2 | AP2 | 47 | 6.6e-15 | 360 | 411 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
TRIUR3_12123-P1 360 SIYRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 411
57****************988532...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.0E-8 | 261 | 317 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.24E-14 | 261 | 327 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 2.52E-17 | 261 | 327 | No hit | No description |
| PROSITE profile | PS51032 | 17.28 | 262 | 325 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.2E-12 | 262 | 326 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.4E-21 | 262 | 331 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-6 | 263 | 274 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.31E-17 | 360 | 420 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.52E-23 | 360 | 419 | No hit | No description |
| Pfam | PF00847 | 8.1E-10 | 360 | 411 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.045 | 361 | 419 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.5E-18 | 361 | 419 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-33 | 361 | 425 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-6 | 401 | 421 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000723 | Biological Process | telomere maintenance | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007389 | Biological Process | pattern specification process | ||||
| GO:0010449 | Biological Process | root meristem growth | ||||
| GO:0019827 | Biological Process | stem cell population maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 600 aa Download sequence Send to blast |
MATVNNWLGF SLSPQELPPS AAASGDVSGA DVCYNIPQDW NMRGSELSAL VAEPKLEDFL 60 GGISYSDHHH HHKQAGGNNM VVPAGSGSGG AACYTTSGGS SVGYLYHPSS ASLQFADSVM 120 VASSGGGVHH DGAGIMANTT ANGDLNNGSG GGGIGLSMIK SWLRSQPSPA QPPEQQRADA 180 AAQGLSLSMN MAACMPPLVG GERGVPELPI VRKDDTAGGS SAGSGAVVSA GGADSTGGSS 240 GVVVETPASR KTADTFGQRT SIYRGVTRHR WTGRYEAHLW DNSCRREGQT RKGRQGGYDK 300 EEKAARAYDL AALKYWGATT TTNFPVNNYE KELEEMKHMT RQEFVASLRR KSSGFSRGAS 360 IYRGVTRHHQ HGRWQARIGR VAGNKDLYLG TFSTQEEAAE AYDIAAIKFR GLNAVTNFDM 420 TRYDVKSILD STALPIGSAA KRLKDAEAAT ASSLALQQQQ HAGVHAQAMQ QQHGGLGSID 480 NAAGASLEHS TGSNSVLYNN GVVESYMLPM STTTTATASH AREQVAAHAD ARAVAAQAYS 540 GGNMSRDHDD GKIAYENYLV STAASGGGRM AAWAQASAPP ATSSSDMTGA QLFSVWNDSN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that promotes cell proliferation, differentiation and morphogenesis, especially during embryogenesis. {ECO:0000269|PubMed:12172019}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00368 | DAP | Transfer from AT3G20840 | Download |
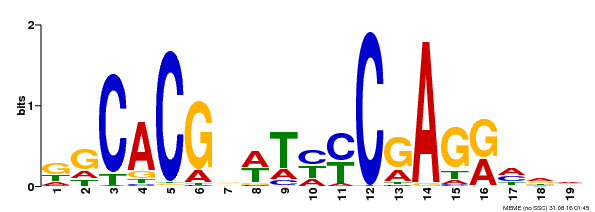
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG670306 | 0.0 | HG670306.1 Triticum aestivum chromosome 3B, genomic scaffold, cultivar Chinese Spring. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020151353.1 | 0.0 | AP2-like ethylene-responsive transcription factor BBM2 | ||||
| Swissprot | Q8LSN2 | 1e-130 | BBM2_BRANA; AP2-like ethylene-responsive transcription factor BBM2 | ||||
| TrEMBL | M7Z7F3 | 0.0 | M7Z7F3_TRIUA; AP2-like ethylene-responsive transcription factor BBM2 | ||||
| STRING | TRIUR3_12123-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1569 | 37 | 110 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17430.1 | 1e-119 | AP2 family protein | ||||



