 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_13265-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 614aa MW: 67084.5 Da PI: 6.9609 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.6 | 9.6e-13 | 460 | 506 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
TRIUR3_13265-P1 460 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYITDLQ 506
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.9E-49 | 56 | 241 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.806 | 456 | 505 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 9.92E-15 | 459 | 510 | No hit | No description |
| SuperFamily | SSF47459 | 1.96E-18 | 459 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.2E-10 | 460 | 506 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.4E-18 | 460 | 518 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-16 | 462 | 511 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 614 aa Download sequence Send to blast |
MEVEEDGANG GNGGAWTEED RDLSTTVLGR DAFAYLTKGG GTISEGLVAA SSPVDLQNKL 60 QELIESEHPG AGWNYAIFWQ LSRTKSGDLV LGWGDGSCRE PNDAELAVAA SAGNDDAKQR 120 MRKRVLQRLH KAFGGADEED YAPTIGQVTD TEMFFLASMY FAFPRRAGAP GQVFAAGLPL 180 WVPNSERNVF PANYCYRGYL ASTAGFRTIL LVPFETGVLE LGSMQQVAES SDTLQTIKSV 240 FAGTGGNKDI IPSREGNGHI ERSPGLAKIF GKDLNLGRSS AGPVIGVSKV DERPWEQRTA 300 GGGSSLLPNV QKGLQSFTWS QARGLNSHQQ KFGNGILIVS NEATHGNNRT ADSSTTTQFQ 360 LQKAPQLQKL PLLQKPPQLV KPLQMVNQQQ LQPQAPRQID FSAGTSSKSG VLVTRAAVLD 420 GDSSEVNGLC KEEGTTPVIE DRRPRKRGRK PANGREEPLN HVEAERQRRE KLNQRFYALR 480 AVVPNISKMD KASLLGDAIA YITDLQKKLK DMETERERFL ESGMVDPRER APRPEVDIQV 540 VQDEVLVRVM SPLENHPVKK VFEAFEEADV RVGESKLTGN NGTVVHSFII KCPGSEQQTR 600 EKVIAAMSRA MSSV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-28 | 454 | 516 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 441 | 449 | RRPRKRGRK |
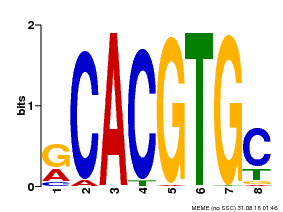
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK376820 | 0.0 | AK376820.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3137B23. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020154206.1 | 0.0 | transcription factor bHLH13-like | ||||
| Refseq | XP_020154207.1 | 0.0 | transcription factor bHLH13-like | ||||
| TrEMBL | M7YZC3 | 0.0 | M7YZC3_TRIUA; Transcription factor bHLH13 | ||||
| STRING | TRIUR3_13265-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 8e-65 | bHLH family protein | ||||



