 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_14996-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 356aa MW: 39608.9 Da PI: 9.9715 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 56.1 | 6.6e-18 | 170 | 216 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ ErrRRdriN+++ L+el+P++ K +Ka+iL +A+eY+ksLq
TRIUR3_14996-P1 170 VHNQSERRRRDRINEKMRSLQELIPHC-----NKADKASILDEAIEYLKSLQ 216
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 6.23E-16 | 161 | 220 | No hit | No description |
| SuperFamily | SSF47459 | 8.37E-21 | 163 | 231 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.177 | 166 | 215 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-21 | 170 | 225 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.9E-15 | 170 | 216 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.8E-17 | 172 | 221 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 356 aa Download sequence Send to blast |
MDLHRSDSDR RAGVSSGDRR KPIWTANSSS SHKMRSHSTD QEISVSDPLP VMSEKAGQEL 60 GNGVEFLGCS PAPALLRFAK KYRRREGRRL VLSLLQWFLL SARWVDRAEN SGGGGAQPRG 120 QSRRPWRHGR QWEIGGESQE ASRDVECEAT EETKSSRRHG SKRRSRAAEV HNQSERRRRD 180 RINEKMRSLQ ELIPHCNKAD KASILDEAIE YLKSLQMQVQ IMWMTTGMAP MMFPGSHQFM 240 PPMAVGMNSA CMPAAQGLNQ MARVPYMNHS LSNHIPMSPS PAMNPMYIAN QMQNIQLREA 300 SNHFLHLDGG QATAPQVAGP YAYTPQVAPK SQIPEVPDCT AVPISGPGQP PAPDGI |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 161 | 178 | KRRSRAAEVHNQSERRRR |
| 2 | 174 | 179 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction (PubMed:17485859). Transcription activator that acts as positive regulator of internode elongation. May function via regulation of cell wall-related genes. May play a role in a drought-associated growth-restriction mechanism in response to drought stress (PubMed:22984180). {ECO:0000269|PubMed:17485859, ECO:0000269|PubMed:22984180}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
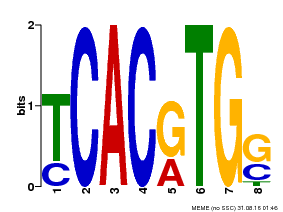
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by light in dark-grown etiolated seedlings (PubMed:17485859). Circadian oscillation under 12 h light/12 h dark cycle conditions, with peaks in the middle of the light period (PubMed:17485859, PubMed:22984180). Down-regulated by cold and drought stresses (PubMed:22984180). {ECO:0000269|PubMed:17485859, ECO:0000269|PubMed:22984180}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK358551 | 0.0 | AK358551.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1078I07. | |||
| GenBank | AK364247 | 0.0 | AK364247.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2023A20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020154183.1 | 1e-152 | transcription factor PIF5-like | ||||
| Refseq | XP_020154184.1 | 1e-152 | transcription factor PIF5-like | ||||
| Swissprot | Q10CH5 | 1e-119 | PIL13_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13 | ||||
| TrEMBL | M7YX84 | 0.0 | M7YX84_TRIUA; Transcription factor PIF5 | ||||
| STRING | TRIUR3_14996-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3174 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 9e-29 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



