 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_21570-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1191aa MW: 133833 Da PI: 7.1305 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.4 | 0.00047 | 1067 | 1090 | 1 | 22 |
EEET..TTTEEESSHHHHHHHHHH CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt 22
++C C++sFs++ +L H r
TRIUR3_21570-P1 1067 FPCDieGCDMSFSSQQDLLLHKRD 1090
789999***************985 PP
| |||||||
| 2 | zf-C2H2 | 10.6 | 0.0018 | 1150 | 1176 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
TRIUR3_21570-P1 1150 YECMvpGCGQTFRFVSDFSRHKRKtgH 1176
788888****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 9.4 | 1067 | 1089 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 15 | 1090 | 1114 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.97 | 1090 | 1119 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.2E-4 | 1092 | 1118 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1092 | 1114 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-10 | 1119 | 1144 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0016 | 1120 | 1144 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.406 | 1120 | 1149 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1122 | 1144 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.42E-10 | 1130 | 1172 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-10 | 1145 | 1173 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.24 | 1150 | 1176 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.076 | 1150 | 1181 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1152 | 1176 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1191 aa Download sequence Send to blast |
MMAFQTLNQK TTVLSPEVLL SAGVPCCRYF SKADWFRTLE NLLSPSLELI IVVLAMVAKE 60 AAIRRASTNC GPMISHYQLL YELALSLSQR EPKDFHTLPR SSRLRDKKKK DEGDTMVKEK 120 FVGSVIENNN LLSILLDKSS CIIVPETPFP PTSFPTMMEP ELTVKQSLTG GHCSISQQAV 180 HNMSVDVALD KSIVVENMSH SQSVTEASLS AYNRRKLYET KYGELGTAAF CLSPSKLQSG 240 VTDKDKRGGL LDQGRLPCVQ CGILSFACVA IIQPREAAVQ FVLSREGISS SAKPGEVSES 300 DGISNWITNN IEMVPQQGQA SEALTVSLAH VSDRCGQLYS RNNNGSTSAL GLLASTYGSS 360 DSEEESNQGK NNQLLETSVS FSSTVQRQRS NSHLYEECCE AKTTTSLLKS IEDNSTTITR 420 CGRDTDINHL AKLREQGTTY DQCSVYVDLA NDLTISGVKA YSDTHVTTAK SSIEPDVLTQ 480 LKYNNDSCRM HVFCLEHALG TWTQLQKIGG ANVMLLCHPE YPRAESAAKF IAEELGLKPD 540 WKDVAFEEAT DDDIRRIQLA LHDEDAEPAS NDWAVKMGIN IYYSAKQSKS PLYSKQIPYN 600 SIIYEAFGQE NPDNLTDYGR QRSGVTKKRV AGCWCGKVWM SNQVHPFLVR EHEEHNRAIV 660 CSKVMLGANY HEIVYDEPSL TCNTMVNCSP SKRISRRKGR DSIEKSGARK KRCSANDEAT 720 LHCSSLGMNS KTISDQPRNF DDHDKHEGGK ITEAPSTQQY QQYTLQSMNM KSSSNKPKDD 780 KGNRNFLDLY DEANDVDCWF NIDSGDNAAI RHLEDSRQQE LGTVKAKSPG KLQGNMRKSS 840 KCKARDDSLN GDKKVQKMNK KSISRKQKDV EINTQFREEY DEDNNWEEVP EGKDHDVKVE 900 SRAKTRSGED DKRNNNSHEL HDEDNDMDCW HDIHGRDNAT MGNLDNSPVQ RLEAAEVKSG 960 GKLHCCKRKS SKGKAKDDYS YGDEKLQMLN IESISRKQKS DSQFDEGCDR DNALDSLLND 1020 VDEAARENCY EVREEEMDGM EVKPRGKMQT GKRKTSRCQA GDKAAKFPCD IEGCDMSFSS 1080 QQDLLLHKRD ICPVKGCKKK FFCHKYLLQH RKVHMDERPL NCEWKGCKKT FKWPWARTEH 1140 MRVHTGVRPY ECMVPGCGQT FRFVSDFSRH KRKTGHSSGV QVLSLKGMLI E |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 6e-56 | 1064 | 1177 | 20 | 133 | Lysine-specific demethylase REF6 |
| 6a58_A | 6e-56 | 1064 | 1177 | 20 | 133 | Lysine-specific demethylase REF6 |
| 6a59_A | 6e-56 | 1064 | 1177 | 20 | 133 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 104 | 110 | RDKKKKD |
| 2 | 695 | 711 | RRKGRDSIEKSGARKKR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
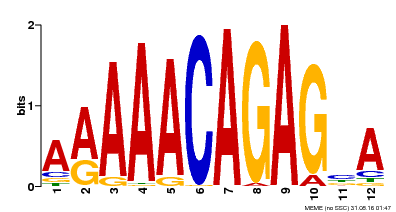
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020201011.1 | 0.0 | lysine-specific demethylase JMJ705-like | ||||
| TrEMBL | T1MUD3 | 0.0 | T1MUD3_TRIUA; Uncharacterized protein | ||||
| STRING | TRIUR3_21570-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6718 | 30 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 6e-93 | relative of early flowering 6 | ||||



