 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_27104-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 589aa MW: 63787.8 Da PI: 4.6842 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.4 | 1.2e-16 | 80 | 127 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +lvd+vk++G g+W+++ + g+ R++k+c++rw ++l
TRIUR3_27104-P1 80 KGPWTSAEDAILVDYVKKHGEGNWNAVQKNTGLFRCGKSCRLRWANHL 127
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 51.9 | 1.8e-16 | 133 | 176 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+eE+ l+++++ ++G++ W++ a++++ gRt++++k++w++
TRIUR3_27104-P1 133 KGAFTPEEERLIIQLHSKMGNK-WARMAAHLP-GRTDNEIKNYWNT 176
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF001693 | 8.4E-258 | 37 | 589 | IPR016310 | Transcription factor, GAMYB |
| PROSITE profile | PS51294 | 16.625 | 75 | 127 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.12E-31 | 77 | 174 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.1E-14 | 79 | 129 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 80 | 127 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-23 | 81 | 134 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.00E-11 | 82 | 127 | No hit | No description |
| PROSITE profile | PS51294 | 25.482 | 128 | 182 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.4E-17 | 132 | 180 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-15 | 133 | 176 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-26 | 135 | 180 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.41E-12 | 135 | 176 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 589 aa Download sequence Send to blast |
MANKTKAVAE SDPRQKQYSV LISDILPYII NEGHDGEMYR VKSESDCEMM HQEDQMDSPV 60 GDDGSSGGSP HRGGGPPLKK GPWTSAEDAI LVDYVKKHGE GNWNAVQKNT GLFRCGKSCR 120 LRWANHLRPN LKKGAFTPEE ERLIIQLHSK MGNKWARMAA HLPGRTDNEI KNYWNTRIKR 180 CQRAGLPIYP ASVCNQSSNE DQQGSSDFNC GENLSSDLLN GNGLYLPDFT CDNFIANSEA 240 LSYAPQLSAV SISSLLGQSF ASKNCGFMDQ VNQAGMLKQP DPLLPGLSDT INGALSSVDQ 300 FSNDSENLKK ALGFDYLHEA NSSSKIIAPF GGALTGSHAF LNGTFSTSRT INGPLKMELP 360 SLQDTESDPN SWLKYTVAPA MQPTELVDPY LQSPTATPSV KSECVSPRNS GLLEELLHEA 420 QGLKSGKNQQ LSVRSSSSSV STPCDTTVVS PEFDICQDYW EEPLNEYAPF SGNSLTGSTA 480 PVSAASPDVF QLSKISPAQS PSLGSGEQAM EPAYEPGAGD TSSHPENFRP DALFSGNTTD 540 SSVFNNAIAM LLGNDMNTDC KPVFGDGIVF DTSPWSNMPH ACQMSEEFK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-32 | 74 | 180 | 1 | 106 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator of gibberellin-dependent alpha-amylase expression in aleurone cells. Involved in pollen and floral organs development. May bind to the 5'-TAACAAA-3' box of alpha-amylase promoter. {ECO:0000269|PubMed:9150608}. | |||||
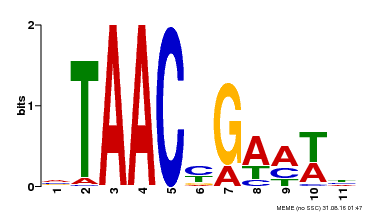
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By gibberellin in aleurone cells. {ECO:0000269|PubMed:9150608}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB214883 | 0.0 | AB214883.1 Triticum monococcum mRNA for transcription factor GAMYB, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020163715.1 | 0.0 | transcription factor GAMYB | ||||
| Refseq | XP_020163716.1 | 0.0 | transcription factor GAMYB | ||||
| Swissprot | A2WW87 | 0.0 | GAM1_ORYSI; Transcription factor GAMYB | ||||
| TrEMBL | M7ZHD6 | 0.0 | M7ZHD6_TRIUA; Transcription factor GAMYB | ||||
| STRING | TRIUR3_27104-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3947 | 36 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.1 | 3e-80 | myb domain protein 33 | ||||



