 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_28079-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 649aa MW: 72002 Da PI: 5.9277 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 402.7 | 4.6e-123 | 39 | 363 | 1 | 311 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraW 94
eel +rmwkd+++lkr+ke++++l ++a+++++k +k ++ a rkkm+raQDgiLkYMlk mevcnaqGfvYgiip+kgkpv+gasd++raW
TRIUR3_28079-P1 39 EELARRMWKDKVRLKRIKEKQQRLA-LEQAELEKSKPKKLSDLALRKKMARAQDGILKYMLKLMEVCNAQGFVYGIIPDKGKPVSGASDNIRAW 131
8*******************99866.55666*************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--T CS
EIN3 95 WkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglsk 188
Wkekv+fd+ngpaai+ky+ +n +l +++s+ +++++ sl++lqD tlgSLLsalmqhc+p+qr++pl+kg++pPWWP+G+e+ww +lgl+k
TRIUR3_28079-P1 132 WKEKVKFDKNGPAAIAKYEVENSLLVNAKSS---GTMNQYSLMDLQDGTLGSLLSALMQHCSPQQRKYPLDKGIPPPWWPSGSEEWWIALGLPK 222
**************************99999...88*********************************************************9 PP
T--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX.XX.XXXX..XXXXXXXXX CS
EIN3 189 dqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah.ss.slrk..qspkvtlsc 278
ppy+kphdlkk+wkv+vL vikh++p++++ir ++r+sk+lqdkm+akes ++l vl++ee ++++ s +l + ++++ +s+
TRIUR3_28079-P1 223 GKT-PPYRKPHDLKKVWKVGVLMGVIKHLAPNFDKIRYHVRKSKCLQDKMTAKESLIWLVVLQREEY-VHSIGNGvSDtHLVDlgDKNECSYSS 314
987.***********************************************************9985.66666664335444455888999999 PP
XXXXXXXXXXXXX.XXXXXXXXXX................XXXXXXXXX CS
EIN3 279 eqkedvegkkeskikhvqavktta................gfpvvrkrk 311
+++dv+ +e+ ++++++++ + + ++kr
TRIUR3_28079-P1 315 CDEYDVDCMEEPPQSTISKDDVGVhqpavhireengsssgNKKHHDKRS 363
99*****999999999999999884444555544444443222222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 9.8E-123 | 39 | 287 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.9E-65 | 161 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 2.88E-58 | 167 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 649 aa Download sequence Send to blast |
MMDNLAIVAK ELGDVSDFEV DGIPNLSEND VSDEEIEAEE LARRMWKDKV RLKRIKEKQQ 60 RLALEQAELE KSKPKKLSDL ALRKKMARAQ DGILKYMLKL MEVCNAQGFV YGIIPDKGKP 120 VSGASDNIRA WWKEKVKFDK NGPAAIAKYE VENSLLVNAK SSGTMNQYSL MDLQDGTLGS 180 LLSALMQHCS PQQRKYPLDK GIPPPWWPSG SEEWWIALGL PKGKTPPYRK PHDLKKVWKV 240 GVLMGVIKHL APNFDKIRYH VRKSKCLQDK MTAKESLIWL VVLQREEYVH SIGNGVSDTH 300 LVDLGDKNEC SYSSCDEYDV DCMEEPPQST ISKDDVGVHQ PAVHIREENG SSSGNKKHHD 360 KRSTQRLPST KETKKPPKRR RHTGQCSVDG SEVEGTQRND NNAEVLSNAI PDMKSNQMEV 420 VCVANPLTSF NPAGTNGGSL QHQGDVQGNF VPPGVVVNNY SQAANIASSS IYMVDQPLAS 480 ESNDYTNSWP GNTFQPDVGL GSIGFSSSSH DYQSSAAKHS VPLSVDNHVP AMGTGALNSS 540 YSHHMAGSGN STSVAGDTQP IMSDAFYIDP DDKFMGSSFD GLPFDFIGIN SPIPDLDELD 600 ELLDDNDLMQ YLGTHQIRKD LTYAIVQCLE PREQPSHARE QETPYMALR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 2e-60 | 167 | 287 | 9 | 129 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
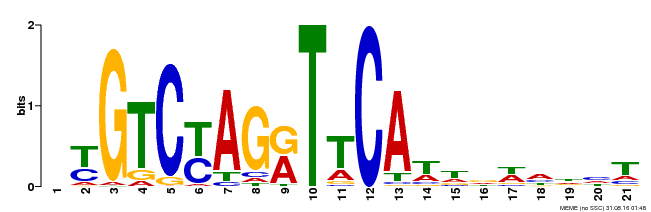
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK371088 | 0.0 | AK371088.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2124J15. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020146451.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_020146452.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_020146453.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_020146454.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_020146456.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_020146457.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | M7ZG52 | 0.0 | M7ZG52_TRIUA; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | TRIUR3_28079-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-121 | ETHYLENE-INSENSITIVE3-like 3 | ||||



