 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_32597-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 857aa MW: 94679.3 Da PI: 6.2314 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 56 | 7.4e-18 | 121 | 184 | 34 | 99 |
SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 34 ktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
++l +d++g++W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F ++++ +l+++++r+
TRIUR3_32597-P1 121 QELFAKDLHGNEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI--WNDNNQLLLGIRRA 184
47999*********************************************..7777778**99997 PP
| |||||||
| 2 | Auxin_resp | 116.4 | 2.5e-38 | 209 | 292 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaast+s F+++YnPras+ eFv++++k++ka++ +++svGmRf+m fete+ss rr++Gt++g+sdld vrWpnS+Wrs+k
TRIUR3_32597-P1 209 AAHAASTNSRFTIFYNPRASPCEFVIPLAKYVKAVYhTRISVGMRFRMLFETEESSVRRYMGTITGISDLDDVRWPNSHWRSVK 292
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01019 | 2.9E-9 | 102 | 185 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.0E-26 | 112 | 198 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.94E-26 | 112 | 212 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 8.9E-16 | 119 | 184 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 9.01E-13 | 121 | 183 | No hit | No description |
| PROSITE profile | PS50863 | 11.166 | 122 | 185 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.0E-33 | 209 | 292 | IPR010525 | Auxin response factor |
| SuperFamily | SSF54277 | 1.18E-7 | 713 | 794 | No hit | No description |
| Pfam | PF02309 | 3.7E-8 | 715 | 804 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 22.381 | 718 | 802 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 857 aa Download sequence Send to blast |
MRLSSPAGAV LPAQAPPSPE VAEEHKCLNS ELWHACAGPL VSLPAVGSRV VYFPQGHSEQ 60 VAASTNKEIE SQIPNYPNLP PQLICQLHNV TMHADAETDE VYAQMTLQPL NPDFTMQPPA 120 QELFAKDLHG NEWKFRHIFR GQPKRHLLTT GWSVFVSAKR LVAGDSVLFI WNDNNQLLLG 180 IRRATRPQTV MPSSVLSSDS MHIGLLAAAA HAASTNSRFT IFYNPRASPC EFVIPLAKYV 240 KAVYHTRISV GMRFRMLFET EESSVRRYMG TITGISDLDD VRWPNSHWRS VKVGWDESTA 300 GERQPRVSLW EIEPLTTFPM YPTPFPLRLK RPWPTGLPSL HGGKDDDLTS SLMWLRDSAN 360 PGFQSLNFGG AGMNPWMQPR LDASLLGLQP DIYQTMAAAA FQDPTKMSPT MLQFQQPQNM 420 VGRATPLLQS QILQQMQPQF QQQSYLQNMN GTTIQGQAQS EFLQQQLQRC QSFNEQKPQL 480 LQQQESNQQQ SQGMQVPQHQ HMQQQKNMAN YQSAYSQLSP GPQSSPTALQ TALPFSQSQS 540 FSDTNMSSLS PSGAPAMHNT LGPFSSETAS HRSMSRPTAV PVPDSWSSKR VAVESLIPSR 600 PQSTTHMEQL SSTPPSIPQS SALAPLPGRG CLVDQDGSSD PQNHLLFGVN IDSHSLLMQG 660 GLHDENDSTA IPYSTSNFLS PSQHDFSLDQ TLNSSGCLDE SGYVPCSHTP DQVNQPPATF 720 VKVYKSGTYG RSLDITKFSN YHELRRELGR LFGLEGQLED PMRSGWQLVF VDREEDVLLV 780 GDDPWQEFVN SVSCIKILSP QEVQQMGKQG LELLSSAPGK RLGGGGGGSS CDDYASRQES 840 RSLSTGIASV GSVEVEF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-126 | 28 | 313 | 51 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
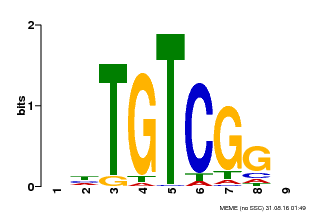
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020157111.1 | 0.0 | auxin response factor 17 | ||||
| Swissprot | Q653U3 | 0.0 | ARFQ_ORYSJ; Auxin response factor 17 | ||||
| TrEMBL | M7YWY8 | 0.0 | M7YWY8_TRIUA; Auxin response factor | ||||
| STRING | TRIUR3_32597-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2012 | 36 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G30330.2 | 0.0 | auxin response factor 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



