 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_33729-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 982aa MW: 107556 Da PI: 6.1134 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 127.9 | 3.9e-40 | 154 | 231 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC adls ak+yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
TRIUR3_33729-P1 154 ACQVEGCCADLSAAKDYHRRHKVCEMHAKANTAVVGNTVQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTRP 231
5**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.6E-33 | 147 | 216 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.291 | 152 | 229 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.88E-37 | 153 | 232 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.1E-29 | 155 | 228 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 4.66E-7 | 762 | 867 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-6 | 764 | 869 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.90E-8 | 767 | 866 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 982 aa Download sequence Send to blast |
MDMEAARVGS QSRHLYGGGL GGELEQARRE KRVFGWDLND WSWDSERFVA TPVPAAVGNG 60 LSLNSSPSSS EEAGAEVSRN GNVRGDFDKR KRVVVIHDDD EKDEGPVGSS NNVLSLRIGG 120 NSVAGGAVED GGVNEEDRNG KKIRVQGGSS SGPACQVEGC CADLSAAKDY HRRHKVCEMH 180 AKANTAVVGN TVQRFCQQCS RFHLLQEFDE GKRSCRRRLA GHNKRRRKTR PENAVGGTPI 240 EEKVSSYLLL SLLGICANLN SDNAEHLQGQ ELLSNLWRNL GNVAKSLDPK ELCKLLETCQ 300 SMQNRSNAGT SEAANALVNT AAAEAAGPSN SKAPFANGGE CGQTSSAVVP VQSNTTMVAA 360 TETPACKFRN FDLNDTCNDV EGFEDGLNCP SWIRQDSTQS PPQTSGNSDS TSAQSLSSSN 420 GDAQCRTDKI VFKLFEKVPS ELPPILRSQI LGWLSSSPTD IESHIRPGCI ILTVYLRLVE 480 SSWRELSENM SVYLDKLSSS SADNFWTSGL VFVMVRRQIA FMHNGSIKLY VNVLLISPYG 540 HITAGQVMLD RPLAPNSHHY CKVLCVSPVA APYSATVNFR VEGFNLVSSS SRLICSIEGR 600 CIFEEDTAIM ADDAEDEDIE YLNFCCSLPD TRGRGFIEVE DSGFSNGFFP FIVAEQNVCS 660 EVCELESTFK SSSLEQPDKD NAMNQALEFL HELGWLLHRV NIISKRDKVE PPVPAFNLLR 720 FRNLGIFAME REWCAVTKML LDLLFDGFVD AGLQSPKEVV LSENLVHSAV RGKSARMVRF 780 LLTYKPNKNL KETAETYLFR PDARGPSAFT PLHIAASTSD AEDVLDALTD DPGLVGLNAW 840 RNARDEIGFT PEDYARQRGN DAYINLVQKK IDRHVGKGHV VLGVPSSMCP GITDGLKAGD 900 ISLEICKAMP MTTTSAARCN ICSRQAKMYP NSFARTFLYR PAMFTVMGVA VICVCVGILL 960 HTLPKVYAAP NFRWELLERG AM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 8e-31 | 148 | 228 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
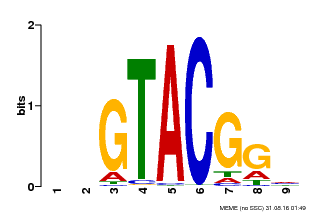
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF447880 | 0.0 | KF447880.1 Triticum aestivum squamosa promoter-binding-like protein 6 (SPL6) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020169994.1 | 0.0 | squamosa promoter-binding-like protein 6 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | M7YUL7 | 0.0 | M7YUL7_TRIUA; Squamosa promoter-binding-like protein 6 | ||||
| STRING | TRIUR3_33729-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



