 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_33948-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 675aa MW: 74455.9 Da PI: 8.5254 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 78.5 | 7e-25 | 119 | 220 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae++ + +++ s++l+ +d++g +W++++iyr++++r++lt+GW+ Fv+ ++L +gD v+F + +
TRIUR3_33948-P1 119 FCKTLTASDTSTHGGFSVPRRAAEDCfppldyS-QQRPSQELVAKDLHGTEWKFRHIYRGQPRRHLLTTGWSAFVNKKKLVSGDAVLFL--RGD 209
99****************************963.3445679************************************************..348 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
++el+++v+r+
TRIUR3_33948-P1 210 DGELRLGVRRA 220
9999***9986 PP
| |||||||
| 2 | Auxin_resp | 113.8 | 1.6e-37 | 245 | 325 | 1 | 82 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsL 82
+aha++tks+F+++YnPr s+seF+v+++k+ k++++++svG+Rfkm++e+ed++err++G+++g+ d+dp W++SkW++L
TRIUR3_33948-P1 245 VAHAVATKSMFQIFYNPRLSQSEFIVPYWKFTKSFSQPFSVGSRFKMRYESEDAAERRYTGIITGTGDADP-MWRGSKWKCL 325
79*********************************************************************.9********9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 2.0E-40 | 113 | 222 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.16E-44 | 114 | 236 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.80E-23 | 117 | 219 | No hit | No description |
| Pfam | PF02362 | 1.0E-22 | 119 | 220 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.3E-23 | 119 | 221 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.31 | 119 | 221 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 4.3E-32 | 245 | 325 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010050 | Biological Process | vegetative phase change | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 675 aa Download sequence Send to blast |
MPRMGGVVVY LPQGHLDHLG DAPAPSPAAV PPHVFCRVVD VTLHADASTA AAVPPHVFCR 60 VVDVTLHADA STDEVYAQLS LLPENEEVVR RMREATEDGS GGEDGETVKQ RFARMPHMFC 120 KTLTASDTST HGGFSVPRRA AEDCFPPLDY SQQRPSQELV AKDLHGTEWK FRHIYRGQPR 180 RHLLTTGWSA FVNKKKLVSG DAVLFLRGDD GELRLGVRRA IRLKNGSAFP ALYSQCSNLG 240 TLANVAHAVA TKSMFQIFYN PRLSQSEFIV PYWKFTKSFS QPFSVGSRFK MRYESEDAAE 300 RRYTGIITGT GDADPMWRGS KWKCLLVRWD DDGEFRRPNR VSPWEIELTS SASGSHLAAP 360 TSKRMKPYLP HANPEFTVPH GGGRPDFAES AQLRKVLQGQ ELLGYRTHDG TAVATSQPCE 420 ARNLQYIDER GCSNNGSNNV LGGVPSHGVR TPLGIPYHCS GFGESQRFQK VLQGQEVFRP 480 YRGSLVDARM RSGGFHQQDG PYASALLDKW RTQQQHAFGF GSSAPVLPSQ PSLSPPSVLM 540 FQQADPKVSR FEFGQGHLDK NMDDPYARFV SAEAIGRGEQ TLSLRPHLGS EVIDTRVAVE 600 NKGVAPTNSC KIFGISLAEK VRARDEMGCD DGGANYPSST QPLKQQVPKS LGNSCATVHE 660 QRPVVGRAID VQQWI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-102 | 1 | 355 | 34 | 361 | Auxin response factor 1 |
| 4ldw_A | 1e-102 | 1 | 355 | 34 | 361 | Auxin response factor 1 |
| 4ldw_B | 1e-102 | 1 | 355 | 34 | 361 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00021 | PBM | Transfer from AT2G33860 | Download |
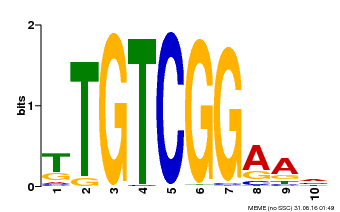
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY376128 | 0.0 | AY376128.1 Triticum aestivum ETTIN-like auxin response factor (ETT1-alpha) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020158279.1 | 0.0 | auxin response factor 15-like | ||||
| Swissprot | Q8S985 | 0.0 | ARFO_ORYSJ; Auxin response factor 15 | ||||
| TrEMBL | M7ZI10 | 0.0 | M7ZI10_TRIUA; Auxin response factor | ||||
| STRING | TRIUR3_33948-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2537 | 35 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G33860.1 | 1e-146 | ARF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



