 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | TRIUR3_34250-P1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 402aa MW: 44583.7 Da PI: 9.3315 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 100.8 | 1.2e-31 | 102 | 193 | 2 | 102 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkelle 95
F+ k+y+++ d++++ l++w + +nsf+v+d f++ +Lp +Fkh nf+SFvRQLn+YgF+kv+ ++ weF+h+sF +g+++ll
TRIUR3_34250-P1 102 FVAKTYQMVCDPRTDALVRWGKGNNSFLVTDVAGFSQLLLPCFFKHGNFSSFVRQLNTYGFRKVHPDR---------WEFAHESFLRGQTHLLP 186
9****************************************************************999.........***************** PP
XXXXXXX CS
HSF_DNA-bind 96 kikrkks 102
+i r+k+
TRIUR3_34250-P1 187 RIVRRKK 193
****987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57756 | 4.71E-9 | 21 | 61 | IPR001878 | Zinc finger, CCHC-type |
| Gene3D | G3DSA:4.10.60.10 | 3.3E-7 | 34 | 56 | IPR001878 | Zinc finger, CCHC-type |
| SMART | SM00343 | 4.6E-5 | 38 | 54 | IPR001878 | Zinc finger, CCHC-type |
| Pfam | PF00098 | 2.8E-7 | 38 | 54 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS50158 | 11.219 | 39 | 54 | IPR001878 | Zinc finger, CCHC-type |
| SMART | SM00415 | 3.3E-49 | 98 | 191 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene3D | G3DSA:1.10.10.10 | 4.0E-34 | 98 | 191 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.36E-31 | 99 | 191 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00447 | 2.2E-27 | 102 | 191 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 7.2E-16 | 102 | 125 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 7.2E-16 | 140 | 152 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 141 | 165 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 7.2E-16 | 153 | 165 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 402 aa Download sequence Send to blast |
MVRRYPKNKN KGKNKSVFNK RTKTTTFKKK KFNKAELECY ACGKPGHFSK ECPERADRRG 60 KTSSKTVNMV TASNTDGMDG LHTELALGLI GCGHGDLQTA PFVAKTYQMV CDPRTDALVR 120 WGKGNNSFLV TDVAGFSQLL LPCFFKHGNF SSFVRQLNTY GFRKVHPDRW EFAHESFLRG 180 QTHLLPRIVR RKKRGEAGAG ASCSSAVGGG EQHQHVVANM GDQVEEEEDE EGREALLEEV 240 QRLRQEQTAI GEQLAKMSRR LQATERRPDR LMSFLSKLAE DPNATSLHLL EQAAEKKRQR 300 MQCPSRDFTS FPVALPLHPA PSPPPPPLLA LGDPTMGGVR VWQWAEPMPL ALTTFEQPSA 360 SSGVQQVPEF EGGRSGSSMG ITDGGTAVET PFPFCLLGQC FF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 7e-21 | 102 | 191 | 20 | 120 | Heat shock factor protein 1 |
| 5d5u_B | 8e-21 | 102 | 191 | 29 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 8e-21 | 102 | 191 | 29 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 8e-21 | 102 | 191 | 29 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00379 | DAP | Transfer from AT3G24520 | Download |
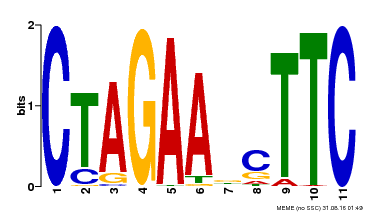
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF208553 | 0.0 | KF208553.1 Triticum aestivum heat shock factor C1e (HsfC1e) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020162679.1 | 0.0 | heat stress transcription factor C-1a-like | ||||
| Swissprot | Q6VBA4 | 1e-124 | HFC1A_ORYSJ; Heat stress transcription factor C-1a | ||||
| TrEMBL | M8AAU0 | 0.0 | M8AAU0_TRIUA; Heat stress transcription factor C-1a | ||||
| STRING | TRIUR3_34250-P1 | 0.0 | (Triticum urartu) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP121 | 37 | 394 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24520.1 | 2e-53 | heat shock transcription factor C1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



