 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG001108t2 | ||||||||
| Common Name | TCM_001108 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 956aa MW: 108076 Da PI: 6.1891 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179 | 5.8e-56 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
++ ++rwl++ ei++iL++++k++++ e+++ p+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+++vl+cyYah+e+n++f
Thecc1EG001108t2 20 MEAQHRWLRPAEICEILKDYKKFHIAPEPAHMPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSIDVLHCYYAHGEDNENF 112
5569***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Lee+l++ivlvhy+evk
Thecc1EG001108t2 113 QRRSYWMLEEDLSHIVLVHYREVK 136
*********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.215 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.9E-77 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 6.9E-49 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 3.8E-6 | 503 | 590 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.01E-18 | 504 | 590 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 5.0E-7 | 504 | 589 | IPR002909 | IPT domain |
| PROSITE profile | PS50297 | 18.112 | 685 | 806 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.21E-13 | 685 | 794 | No hit | No description |
| SuperFamily | SSF48403 | 7.31E-17 | 686 | 796 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.6E-17 | 691 | 800 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 5.2E-7 | 707 | 799 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0046 | 735 | 764 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.912 | 735 | 767 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 490 | 774 | 803 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 8.956 | 911 | 939 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 956 aa Download sequence Send to blast |
MAETRRYGLS NQLDIEQILM EAQHRWLRPA EICEILKDYK KFHIAPEPAH MPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKAGSIDVLH CYYAHGEDNE NFQRRSYWML 120 EEDLSHIVLV HYREVKGNRT NFNRIKETEE AIPYSQDTEG ILPNSEMESS VSSSFHPNNG 180 QIPSKTTDTT SLNSVQASEY EDAESDYNHQ ASSQFNSFLE LQQPVVGRVD SGFSDPYVPL 240 SHSNDYHGKP SGTGFQLTQP DKSREYNDAG LTYEPQKNLD FTSWEDVLEN CTPGVESAQH 300 QPPFSSTQRD TMGQLFNNSF LTKQEFDNQA PVQEEWQASE GDSSHLSKWP LNQKLHPDLR 360 YDLTFRFHEQ EVNHHVHPDK QHDNSMQNNE QIEPSNGKHG YALKPDPESH LTLEGKSINS 420 SAMRQHLFDG SLVEEGLKKL DSFNRWMSKE LGDVDESHMQ SSSGAYWDAV EGQNGVDVST 480 IPSQGQLDTF LLGPSLSQDQ LFSIIDFSPN WAYVGSEIKV LITGRFLKSR DEAENCKWSC 540 MFGEVEVPAE VIADGVLRCH TPIHKAGRVP FYVTCSNRLA CSEVREFEYR VNHMETMDYP 600 RSNTNEILDM RFGRLLCLGP RSPYSITYNV ADVSQLSDEI NSLLKEDIKE WDQMLMHNSA 660 EEISPEKMKE QLLQKLLKEK LRVWLLQKVA EGGKGPNILD DGGQGVIHFA AALGYDWALE 720 PTIVAGVSVN FRDVNGWTAL HWAASYGRER TVASLISLGA APGALTDPTP KYPLGRTPAD 780 LASTNGHKGI SGYLAESDLS FHLRSLNLDN QGNNDTVDSR ADAIQKILER STAPLGCGDA 840 SDGPSLKDSL AAVRNATQAA ARIHQVFRVQ SFQKRQLKEY GDGKFGMSNE RALSLIAVKS 900 NKPGQHDEHV QAAAIRIQNK FRGWKGRKEF LIIRQRIRSC TIRFSAVQKQ FWMKT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cxk_A | 5e-13 | 504 | 590 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_B | 5e-13 | 504 | 590 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_C | 5e-13 | 504 | 590 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_D | 5e-13 | 504 | 590 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_E | 5e-13 | 504 | 590 | 9 | 89 | calmodulin binding transcription activator 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
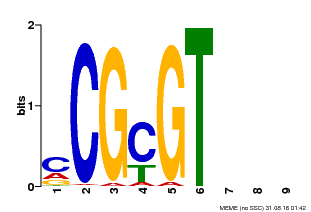
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007047945.2 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A061DQ83 | 0.0 | A0A061DQ83_THECC; Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains, putative isoform 2 | ||||
| STRING | EOX92102 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG001108t2 |
| Entrez Gene | 18611563 |



