 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG004922t2 | ||||||||
| Common Name | TCM_004922 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 983aa MW: 108368 Da PI: 6.4526 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.1 | 3.3e-40 | 149 | 226 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C adls k+yhrrhkvCe+hska+++lv +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Thecc1EG004922t2 149 VCQVEDCGADLSCSKDYHRRHKVCEMHSKASKALVGNVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 226
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.3E-33 | 142 | 211 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.829 | 147 | 224 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.22E-37 | 148 | 230 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.0E-29 | 150 | 223 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 2.35E-6 | 753 | 865 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-5 | 753 | 878 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.64E-6 | 754 | 865 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 983 aa Download sequence Send to blast |
MEARFGSDAH HFYGMNPANL RAVGKRTLEW DLNDWKWDGD LFIASSINPV SADSTGRQFF 60 PLGSGIPGNS SNSSSSCSDE VNLETEKGKR ELEKKRRVIV VEDDSPNEEA GSLTLKLGGQ 120 GGHGYPISQR EGTSGKKTKL GGGSGNRAVC QVEDCGADLS CSKDYHRRHK VCEMHSKASK 180 ALVGNVMQRF CQQCSRFHVL QEFDEGKRSC RRRLAGHNKR RRKTNPDTVV NGNSLNDEQT 240 SGYLLLSLLK ILSNMHSNRS DQTTDQDVLS HLLRSLANHT GEQGGRNISG LLPEPQDSEA 300 VSALFLNGQG PPRPFKQHHT GAASEMAEKG VSSQGTRGVK VQGNTAGAVK MNNFDLNDIY 360 IDSDEGTDDI ERSPAAVNTG TSSLDCPSWI QQDSHQSSPP QTSGNSDSAS AQSPSSSSGD 420 AQQSRTDRIV FKLFGKEPND FPMVLRAQIL DWLSHSPTDI ESYIRPGCIV LTIYLRQAEA 480 AWDELCCDLS FTLSRLLDCS DDTFWRSGWI YIRVQDQIAF IYNGQVVVDT SLPLRSNHYS 540 KITSVKPIAI SATERAQFSV KGINLSRPAT RLLCAVEGKC LLQETTNELM DGNDDYKEQD 600 ELQCVNFSCS VPTVTGRGFI EIEDHGFSSS FFPFIVAEED VCSEVRMLES VLEISDTDAD 660 VGGTGKLEAK HRAMDFIHEV GWLLHRCQLK SRLGHLDPNP EPFPLSRFKW LMEFSMDHEW 720 CAVVKKLLNI LLNGVVGSGE HPSLNLALTE MGLLHRAVRK NCRPLVELLL RFVPEKASDK 780 LGFENETLTG VDHKSFLFRP DVLGPAGLTP LHIAAGKDGS EDVLDALTDD PGKVGIDAWK 840 SARDSTGSTP EDYARLRGHY SYIHLVQKKI NKRTASGHVV VDIPGALSEC SMNQKQNNES 900 TSSFEIGRLE LRSIQRHCKL CDQKLAYGCG TTSKSLVYRP AMLSMVAIAA VCVCVALLFK 960 SCPEVLYVFR PFRWELLDYG TS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-30 | 142 | 223 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
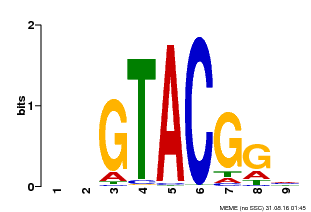
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ622325 | 0.0 | KJ622325.1 Gossypium hirsutum SQUAMOSA promoter binding-like transcription factor (SPL17) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017979528.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A061DRR7 | 0.0 | A0A061DRR7_THECC; Squamosa promoter-binding protein, putative isoform 2 | ||||
| STRING | EOX95415 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG004922t2 |
| Entrez Gene | 18613790 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



