 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG007482t1 | ||||||||
| Common Name | TCM_007482 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1065aa MW: 119670 Da PI: 5.8539 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 184.9 | 8.8e-58 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
l+ ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+een++f
Thecc1EG007482t1 20 LEAQHRWLRPAEICEILRNYQKFHISSEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEENENF 112
5679***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Le+el +iv+vhylevk
Thecc1EG007482t1 113 QRRSYWMLEQELMHIVFVHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.873 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-83 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.7E-51 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 3.7E-6 | 468 | 580 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 6.02E-17 | 494 | 580 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 2.0E-5 | 494 | 573 | IPR002909 | IPT domain |
| CDD | cd00204 | 6.58E-16 | 676 | 785 | No hit | No description |
| Pfam | PF12796 | 1.2E-6 | 676 | 754 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.359 | 676 | 787 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.35E-18 | 677 | 787 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.8E-19 | 679 | 789 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0026 | 726 | 755 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.018 | 726 | 758 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2000 | 765 | 794 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.37 | 887 | 909 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 1.67E-7 | 887 | 938 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 8.096 | 888 | 917 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0017 | 889 | 908 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 910 | 932 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.413 | 911 | 935 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 913 | 932 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1065 aa Download sequence Send to blast |
MADRASYSLA PRLDIEQILL EAQHRWLRPA EICEILRNYQ KFHISSEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEENE NFQRRSYWML 120 EQELMHIVFV HYLEVKGSRT IGGIRDTGDV SNSQTSSPST SSYSVSHTKA PSGNTDSASP 180 TSTLTSLCED ADSEDSHQAS SRIPTSPQVG NATMMDKMDP GFLNPYSSHP FPGRSSIPGV 240 NEVSHLHGDR PMGIDYGTYM TEAQKTLDLA SWEGGLEQYM PLYPVVSSHA SMASAQPDTM 300 SISQQQMMKG KQLDVESADK EFGNLLPTQS NWQIPLADNA LELPKWPMDQ SSNFELAYDT 360 RLFEQKTDDF HLPNALEEFT NNDVLNEQPV HKNLQTQLIN ADTNSVMKSY PENDTHLEGN 420 INYAFSLKKS LLDGEESLKK VDSFSRWITK ELGEVDNLQM QSSSGIAWSS VECGNVSDDA 480 SLSPSISQDQ LFSIVDFSPK WAYTDLETEV LIIGTFLKSQ EEVAKYNWSC MFGEVEVPAE 540 VIADGILFCH APPHSVGQVP FYVTCSNRLA CSEVREFDYR AGFAKGIHVS HIYGVASTEM 600 LLRFQMLLSL KSFSSLNHHL EGVGEKRDLI AKIILMKEEE ECHQIVDPSS DKDLSQREEK 660 EWLLQKLMKE KLYSWLLHKI VEDGKGPNIL DEKGQGVLHL AAALGYDWAL KPTVTAGVSI 720 NFRDVNGWTA LHWAAFCGRE QTVAILVFLG ADPGALTDPS PEFPLGRTPA DLASDNGHKG 780 ISGFLAESSL TSYLSSLTMN DAKAAVQTVS ERMATPVNDS DLQDILLKDS ITAVCNATQA 840 ADRIHQMFRL QSFQRKQLTE SGDAVSDEHA ISIVTAKARR SLQSEGVAHA AATQIQKKFR 900 GWKKRKEFLL IRQRIVKIQA HVRGHQVRKQ YRTIIWSVGI LEKVILRWRR KGSGLRGFRR 960 DALTKEPESQ CMPTKEDEYD FLKEGRKQTE ERLQKALTRV KSMAQNPEGR GQYRRLLTLV 1020 QGIRENKACN MVMNSTEEVA DGDEDLIDID SLLDDDNFMS IAFE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ydw_A | 4e-11 | 680 | 785 | 66 | 165 | DARPIN 44C12V5 |
| 4ydw_B | 4e-11 | 680 | 785 | 66 | 165 | DARPIN 44C12V5 |
| 4ydy_A | 4e-11 | 680 | 785 | 66 | 165 | DARPIN 44C12V5 |
| 4ydy_B | 4e-11 | 680 | 785 | 66 | 165 | DARPIN 44C12V5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
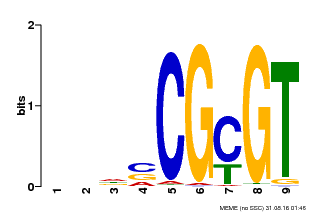
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN558810 | 9e-74 | JN558810.1 Solanum lycopersicum calmodulin-binding transcription factor SR1L mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007042960.2 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A061E376 | 0.0 | A0A061E376_THECC; Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains, putative isoform 1 | ||||
| STRING | EOX98791 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG007482t1 |
| Entrez Gene | 18608292 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



