 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG011212t1 | ||||||||
| Common Name | TCM_011212 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 283aa MW: 31523.2 Da PI: 10.2867 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 346.9 | 4.8e-106 | 1 | 282 | 1 | 301 |
GAGA_bind 1 mdddgsrernkgyyepaaslkenlglqlmssiaerdaki....rernlalsekkaavaerd.........maflqrdkalaernkalverdnk 80
mddd+ + rn+gyyep+ +k +lglqlmss+aerd+k r+ nl + +++aa+++rd m++ rd+++++ ++k
Thecc1EG011212t1 1 MDDDALNMRNWGYYEPS--FKGHLGLQLMSSMAERDTKSfipgRDPNL-MVTPNAAFHPRDcvvseapipMNY-VRDSWISQ--------REK 81
9***9999*******99..*******************99********.************************.9*******........789 PP
GAGA_bind 81 llalllvenslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksek 173
++++l+++ ++++++l++t+ ++sl l++++ + +++e + +e +++++ke kkrq ++ pk++kakk++k
Thecc1EG011212t1 82 FFNMLPAT-------APNYGILPETSAAHSLPI-----LQPPPDPSSRNER-VVGRVE-EQSVNKEGVPLKKRQGGAAPKTPKAKKPRK---- 156
*****954.......4689**********9988.....8888888544443.333444.455666666777799999999999999999.... PP
GAGA_bind 174 skkkvkkesaderskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafk 266
+k+++++ +r+k kks+d+ +ng ++D+s++P+PvCsCtG+++qCY+WG+GGWqSaCCtt++S+yPLP+stkrrgaRiagrKmSqgafk
Thecc1EG011212t1 157 PKDNTNSTV--QRVKPAKKSMDIKINGYDMDISGIPIPVCSCTGSPQQCYRWGCGGWQSACCTTNVSMYPLPMSTKRRGARIAGRKMSQGAFK 247
333334444..689******************************************************************************* PP
GAGA_bind 267 klLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
k+LekLaae y+++np+DL++hWA+HGtnkfvtir
Thecc1EG011212t1 248 KVLEKLAAENYNFNNPIDLRTHWARHGTNKFVTIR 282
**********************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 3.3E-114 | 1 | 282 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 3.5E-174 | 1 | 282 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MDDDALNMRN WGYYEPSFKG HLGLQLMSSM AERDTKSFIP GRDPNLMVTP NAAFHPRDCV 60 VSEAPIPMNY VRDSWISQRE KFFNMLPATA PNYGILPETS AAHSLPILQP PPDPSSRNER 120 VVGRVEEQSV NKEGVPLKKR QGGAAPKTPK AKKPRKPKDN TNSTVQRVKP AKKSMDIKIN 180 GYDMDISGIP IPVCSCTGSP QQCYRWGCGG WQSACCTTNV SMYPLPMSTK RRGARIAGRK 240 MSQGAFKKVL EKLAAENYNF NNPIDLRTHW ARHGTNKFVT IR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
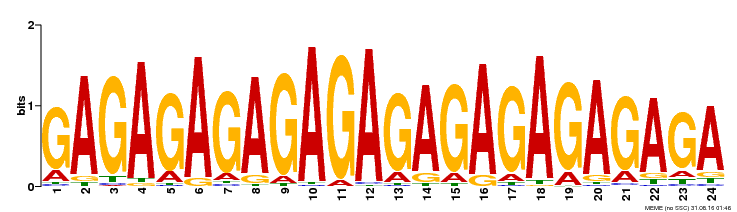
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007045434.2 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_007045435.2 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE2 | ||||
| Refseq | XP_007045436.2 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE2 | ||||
| Swissprot | Q9SKD0 | 1e-111 | BPC1_ARATH; Protein BASIC PENTACYSTEINE1 | ||||
| TrEMBL | A0A061E9H4 | 0.0 | A0A061E9H4_THECC; Basic pentacysteine 2 isoform 1 | ||||
| STRING | EOY01266 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2893 | 27 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14685.3 | 1e-111 | basic pentacysteine 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG011212t1 |
| Entrez Gene | 18609982 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



