 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG011217t1 | ||||||||
| Common Name | TCM_011217 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 184aa MW: 21064.1 Da PI: 10.8636 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 74.5 | 1.6e-23 | 3 | 52 | 3 | 57 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrev 57
k +Y++C++NhA lGg+a+DGC Ef+ps+++ +C ACgCHRn+HR+ +
Thecc1EG011217t1 3 KETYRDCQRNHACYLGGYAIDGCSEFIPSNNK-----ESHCKACGCHRNYHRKVP 52
679*************************9544.....579************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04770 | 3.0E-19 | 5 | 51 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 9.8E-18 | 5 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| ProDom | PD125774 | 5.0E-10 | 6 | 52 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 21.9 | 6 | 51 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-14 | 98 | 163 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.53E-6 | 100 | 162 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 1.2E-22 | 106 | 163 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 184 aa Download sequence Send to blast |
MEKETYRDCQ RNHACYLGGY AIDGCSEFIP SNNKESHCKA CGCHRNYHRK VPFIYSKEYP 60 EIGHRSGKSF WSLREAKRIA RQHRVLPAPS PQVLVLKQQN SNRKFKQRKS KFTGGQREAM 120 RALAESLGWT MRNKGRQTEI NRFCGRIGVS RLHFKTWLNN NKKLYFKGTA SMSGANSSSS 180 KAV* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 101 | 108 | NRKFKQRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00143 | DAP | Transfer from AT1G14687 | Download |
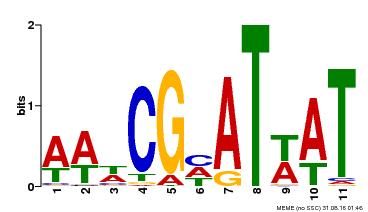
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007045440.2 | 1e-133 | PREDICTED: zinc-finger homeodomain protein 14 | ||||
| TrEMBL | A0A061EA71 | 1e-135 | A0A061EA71_THECC; Homeobox protein 32, putative | ||||
| STRING | EOY01272 | 1e-135 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13728 | 21 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14687.1 | 1e-38 | homeobox protein 32 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG011217t1 |
| Entrez Gene | 18609986 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



