 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG029792t2 | ||||||||
| Common Name | TCM_029792 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 607aa MW: 66897.4 Da PI: 9.8552 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 32.4 | 1.6e-10 | 72 | 115 | 54 | 99 |
EETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 54 kksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
++++r++lt+GW+ Fv+ ++L +gD v+F + +++el+++++r+
Thecc1EG029792t2 72 GQPRRHLLTTGWSAFVNKKKLVSGDAVLFL--RGEDGELRLGIRRA 115
67899*************************..449999*****996 PP
| |||||||
| 2 | Auxin_resp | 119.1 | 3.5e-39 | 140 | 222 | 1 | 82 |
Auxin_resp 1 aahaastksvFevvYnP.rastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsL 82
++ha+s+ksvF+++YnP ras+seF+++v+k+ k+l+++++vGmRfkm+feted++err++G+v+g+sd+dpvrWp+SkWr+L
Thecc1EG029792t2 140 VVHAISMKSVFSIYYNPsRASSSEFIIPVHKFWKSLDHSFAVGMRFKMRFETEDAAERRYTGLVTGISDMDPVRWPGSKWRCL 222
689**************7799*************************************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 8.37E-9 | 19 | 115 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 9.545 | 62 | 116 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.4E-8 | 71 | 115 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.5E-12 | 71 | 118 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF06507 | 9.6E-34 | 140 | 222 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010050 | Biological Process | vegetative phase change | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 607 aa Download sequence Send to blast |
MSKQISSQRR PTCSARPLRL LIPVHMEASL FRVELLRTAS RPWIIISRGP HKSLLQKTFM 60 AWNGGFDTFI GGQPRRHLLT TGWSAFVNKK KLVSGDAVLF LRGEDGELRL GIRRAAQIKN 120 GTSFPSLCSQ QLNRSNFADV VHAISMKSVF SIYYNPSRAS SSEFIIPVHK FWKSLDHSFA 180 VGMRFKMRFE TEDAAERRYT GLVTGISDMD PVRWPGSKWR CLLVRWDDID ANRHSRVSPW 240 EIELSGTISS SNNLLTPGVK RNRIGLPSGK PEFMVPDGIG ASDFGEPLRF QKVLQGQEIL 300 GFNTLYDGAD GQNLHRSEIR RCFPGSNGSG IVAIGNVGRD PLGNSEISYR GVGFGESFRF 360 HKVLQGQEIY VSPPYIRGPT TNDTQENDGL GVRDAGQFSG TRSGWSSLMQ SYNTHSHIRP 420 SAPSAQVSSP SSVLMFQQAS NPIPNINPIY SVNNQEKEQG VDNRSSFRAP EMYGGKLLQS 480 STGECSSRGR HHGTFDSFGH SNDSVQLGDS QPLAAQPTFR TSQDIASSCK SSCRLFGFSL 540 TEGSQDANKE DNMVKATSSL GPRAFLPCIG EQFHPKPPAV TNTVGSNCTK VSNLYAVRDM 600 LFDIAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldy_A | 2e-47 | 66 | 251 | 176 | 361 | Auxin response factor 1 |
| 4ldy_B | 2e-47 | 66 | 251 | 176 | 361 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
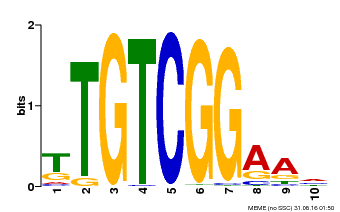
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00021 | PBM | Transfer from AT2G33860 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017978888.1 | 0.0 | PREDICTED: auxin response factor 3 isoform X1 | ||||
| Swissprot | Q8S985 | 1e-113 | ARFO_ORYSJ; Auxin response factor 15 | ||||
| TrEMBL | A0A061GF00 | 0.0 | A0A061GF00_THECC; Auxin response factor | ||||
| STRING | EOY28143 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G33860.1 | 1e-109 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG029792t2 |
| Entrez Gene | 18596775 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



