 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG034649t2 | ||||||||
| Common Name | TCM_034649 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 399aa MW: 43408.4 Da PI: 6.3215 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 31.2 | 3.8e-10 | 239 | 287 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L+el+P + +k Ka +L + ++Y++sLq
Thecc1EG034649t2 239 NSHSLAERVRREKISERMRLLQELVPGC----NKITGKAVMLDEIINYVQSLQ 287
58**************************....999*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.96E-11 | 234 | 291 | No hit | No description |
| SuperFamily | SSF47459 | 6.02E-17 | 234 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.485 | 236 | 286 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-16 | 237 | 304 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.4E-7 | 239 | 287 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.6E-10 | 242 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
MAMSSISMYN KPSNASDPFF GSSWDPIVSL SQSESLVGSS MVSHSEFANS HYPLLMENQG 60 ITSTAHFSQY QSDPSFVELV PKLPGFGSGN WSEMVGPFSL PQCGQIANGK CPQNYALNTE 120 VGNERACTNS TQSRDEHQLS DEGVVGASPN GKRRKRVPES NSPLRSYQNA DEEPQKDPSG 180 ESSDVPKGQD EKIQKTEQIT GVNSRGKQIA KQAKDSSQTG EAPKENYIHV RARRGQATNS 240 HSLAERVRRE KISERMRLLQ ELVPGCNKIT GKAVMLDEII NYVQSLQQQV EFLSMKLATV 300 NPELNIDLER VLSKDILHSR GGSAGILGFG PGLNSSHPFP PRIFPGTISG VPSTNPQFPP 360 LPQTVLDNEL QNLFQMGYDS SSAMDSLGPN AGRLKSEL* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00135 | DAP | Transfer from AT1G10120 | Download |
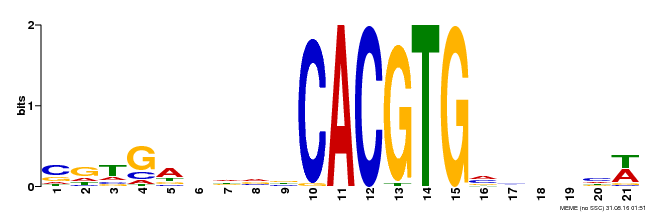
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007018426.2 | 0.0 | PREDICTED: transcription factor bHLH74 isoform X1 | ||||
| TrEMBL | A0A061FF54 | 0.0 | A0A061FF54_THECC; Basic helix-loop-helix DNA-binding superfamily protein, putative isoform 2 | ||||
| STRING | EOY15650 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G10120.1 | 1e-99 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG034649t2 |
| Entrez Gene | 18591929 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



