 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thecc1EG040510t2 | ||||||||
| Common Name | TCM_040510 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Theobroma
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 599aa MW: 67016.8 Da PI: 5.2112 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 428.7 | 5.7e-131 | 38 | 364 | 1 | 303 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLra 93
eel++rmwkd+++l+r+ker+k +++ a++++k +++++qarrkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gas+++ra
Thecc1EG040510t2 38 EELERRMWKDRIKLNRIKERQKIA--AQQ-AAEKQKPKQTTDQARRKKMSRAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASENIRA 127
8**************999999963..455.599************************************************************ PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT- CS
EIN3 94 WWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelgl 186
WWkekv+fd+ngpaai+ky+a+++++s+++s+ ++ + ++sl++lqD+tlgSLLs+lmqhcdppqr++plek ++pPWWPtG+e+ww +lgl
Thecc1EG040510t2 128 WWKEKVKFDKNGPAAIAKYEAECIAMSESDSN--RNGNLQSSLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKRIPPPWWPTGNEEWWVRLGL 218
*********************99999987777..689999***************************************************** PP
-TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.........XXXXX CS
EIN3 187 skdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.........slrkq 270
++ q ppykkphdlkk+wkv+vLtavikhmsp+i++ir+l+rqsk+lqdkm+akes+++l vl++ee+++++ s+++ r +
Thecc1EG040510t2 219 QQGQT-PPYKKPHDLKKMWKVGVLTAVIKHMSPDIAKIRRLVRQSKCLQDKMTAKESAIWLGVLSREEALIQQPSSDNGtsgitemlaGGRGN 310
*9987.***********************************************************************4445688777655669 PP
XXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX.....................X CS
EIN3 271 spkvtlsceqkedvegkkeskikhvqavktta.....................g 303
+++ +s+++++dv+g +e + ++++++++ +
Thecc1EG040510t2 311 KKQPAISSDSDYDVDGGEEGVGSVSSKDDRRNqptdaepvayisndvshpvqeK 364
99999**********888887787777777777788888888888665555442 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.5E-124 | 38 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 3.5E-71 | 159 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.96E-61 | 166 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 599 aa Download sequence Send to blast |
MGEFEDIGAD ISSDIEVDDL RCENIAEKDV SDEEIEAEEL ERRMWKDRIK LNRIKERQKI 60 AAQQAAEKQK PKQTTDQARR KKMSRAQDGI LKYMLKLMEV CKARGFVYGI IPEKGKPVSG 120 ASENIRAWWK EKVKFDKNGP AAIAKYEAEC IAMSESDSNR NGNLQSSLQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKRI PPPWWPTGNE EWWVRLGLQQ GQTPPYKKPH DLKKMWKVGV 240 LTAVIKHMSP DIAKIRRLVR QSKCLQDKMT AKESAIWLGV LSREEALIQQ PSSDNGTSGI 300 TEMLAGGRGN KKQPAISSDS DYDVDGGEEG VGSVSSKDDR RNQPTDAEPV AYISNDVSHP 360 VQEKEPVGKH PKKRRRLRSS DGDEQPGPSD EHANTESRST LPDINQTDVS FLEYQMPGAQ 420 KENDASTALR HVEKGLDVPP HLPASGYSHY PAIPSANKIS TQSMYVDGRP MLYPLVQDAE 480 LPNGAAYEFY NPSVEFGPTH EGQQTQMEMN VTQIRPENGV HVSLPGRNEN EITVGELHHY 540 VKDIFQNEQD RAGHNPFGSP TSDPSLYGAF HSPYEFGFDG TSSLDELFDD DLIEYFGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-72 | 165 | 296 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 370 | 376 | PKKRRRL |
| 2 | 372 | 376 | KRRRL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
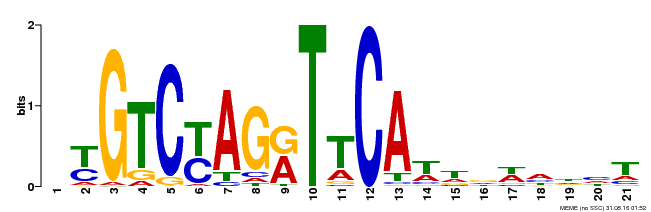
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017982625.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_017982626.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A061GSY6 | 0.0 | A0A061GSY6_THECC; Ethylene-insensitive 3f isoform 1 | ||||
| STRING | EOY32534 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-179 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thecc1EG040510t2 |
| Entrez Gene | 18589744 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



