 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10000426m | ||||||||
| Common Name | EUTSA_v10000426mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 299aa MW: 33506.5 Da PI: 8.3297 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.7 | 1.8e-12 | 119 | 166 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+ h +Er+RR+++ ++f L l+P + kK++Ka++L A+++Ik+Lq
Thhalv10000426m 119 QDHILAERKRREKLSQRFVALSALIPGL-----KKMDKASVLGDAIKHIKNLQ 166
56999***********************.....9******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.618 | 116 | 165 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.5E-10 | 120 | 166 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-16 | 120 | 182 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.02E-17 | 120 | 185 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 9.72E-13 | 121 | 170 | No hit | No description |
| SMART | SM00353 | 1.2E-14 | 122 | 171 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 299 aa Download sequence Send to blast |
MNSLVGDVPQ SLSSLDDITS CYNFDSSCND DLIEKKPAKI VKTTHISPKL HHFPSSNPLP 60 PKLQPSSRIL SFEKMGFKNM NQNSPNLIFS AKEEAGSPEL ITRETKRAQP LTRSPSNSQD 120 HILAERKRRE KLSQRFVALS ALIPGLKKMD KASVLGDAIK HIKNLQESVK EFEEQKKERT 180 MESMVLVKKS QFVLNDYDQP PSSSSDGNHE FSSLNLPEME VRVSGKEVLI KMLCDKQKGH 240 VIKIMDEIEK LDLSITNSIV LPFGPTFDIT IIAQKNNDFD MKIEDVTKSL SCGLSKFM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-13 | 120 | 176 | 8 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 124 | 129 | ERKRRE |
| 2 | 125 | 130 | RKRREK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00273 | DAP | Transfer from AT2G22750 | Download |
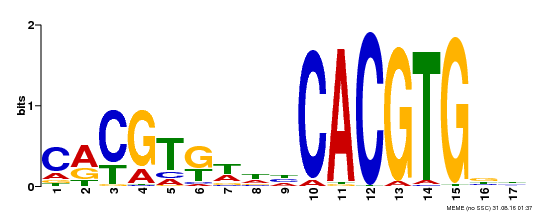
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10000426m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ446549 | 0.0 | DQ446549.1 Arabidopsis thaliana clone pENTR221-At2g22750 basic helix-loop-helix family protein (At2g22750) mRNA, complete cds. | |||
| GenBank | DQ653015 | 0.0 | DQ653015.1 Arabidopsis thaliana clone 0000017319_0000012121 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006404738.2 | 0.0 | transcription factor bHLH18 | ||||
| Swissprot | Q1PF17 | 1e-138 | BH018_ARATH; Transcription factor bHLH18 | ||||
| TrEMBL | V4NIV1 | 0.0 | V4NIV1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006404738.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM492 | 27 | 146 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22750.2 | 1e-141 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10000426m |
| Entrez Gene | 18021788 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



