 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10001222m | ||||||||
| Common Name | EUTSA_v10001222mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 674aa MW: 75444.7 Da PI: 6.243 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 157.7 | 8e-49 | 80 | 228 | 1 | 144 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspessl 94
gg+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+V++AL+reAGw v++DGttyr++ +p+ +++++ ++s+es+l
Thhalv10001222m 80 GGKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVIAALAREAGWSVDADGTTYRQSHQPN----HVVQFPTRSIESPL 169
6899***********************************************************************9....999**********9 PP
DUF822 95 q.sslkssalaspvesysaspksssfpspssldsislasa........asllpvlsvls 144
+ s+lk++++a+ +++++++ ++ +sp+sl s+ +a++ + +p+ sv +
Thhalv10001222m 170 SsSTLKNCTKATLDCQQHPVLRIEENLSPVSLGSVVIAENdhpgngryTGASPIASVGC 228
9899********************************99987777666655566666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 3.1E-46 | 81 | 225 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.16E-154 | 248 | 674 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 2.8E-166 | 251 | 674 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 5.6E-82 | 256 | 649 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 287 | 301 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 308 | 326 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 330 | 351 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 418 | 440 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 491 | 510 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 525 | 541 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 542 | 553 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 560 | 583 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.6E-49 | 598 | 620 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 674 aa Download sequence Send to blast |
MHTLNTTGSR DPINPNPNPN SNLDPDQFPH RNNNHPQSRR PRGFAAAAAA SMCPAESDNG 60 NIAGIGGGET SSGGGGGGGG GKGKREREKE KERTKLRERH RRAITSRMLA GLRQYGNFPL 120 PARADMNDVI AALAREAGWS VDADGTTYRQ SHQPNHVVQF PTRSIESPLS SSTLKNCTKA 180 TLDCQQHPVL RIEENLSPVS LGSVVIAEND HPGNGRYTGA SPIASVGCVE ANQLIQDVHS 240 TEPRTGFTES FYVPVYAMLP VGIIDNFGQL GDPVGVKQEL SYMKSLNVDG VVIDCWWGIV 300 EGWNPQKYVW SGYKELFNLI REFKLKLQVV MAFHEYGGNA VSLPQWVLEI GKDNPDIFFT 360 DREGRRNFEC LNWCIDKERV LHGRTGIEVY FDFMRSFRSE FDDFFEEGLI TAIEIGLGAS 420 GELKYPSFPE RMGWIYPGIG EFQCYDKYSQ TSLLKEAKSR GFAFWGKGPE NAGQYNSQPH 480 ETGFFQERGE YDSYYGRFFL NWYSQLLIGH AENVLSLANL AFEETKIIVK IPAIYWSYKT 540 ASHAAELTAG YYNPSNRDGY SLLFETLKKY SVTVKFVCPG QLMSTNEHEE GLADPEGLSW 600 QVINAAWDKG LLIGGENAIT CFDREGCMRL IEMAKPRNHP DSCHFSFFTY RQPSPLVQGS 660 SCFADLDYFI KRMH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-118 | 252 | 674 | 11 | 442 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
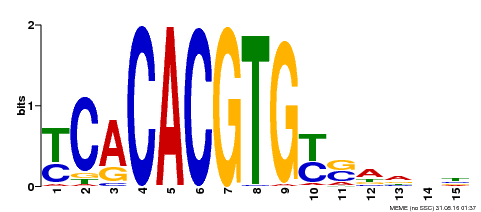
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10001222m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK117140 | 0.0 | AK117140.1 Arabidopsis thaliana At5g45300 mRNA for putative beta-amylase, complete cds, clone: RAFL16-68-D16. | |||
| GenBank | BT006482 | 0.0 | BT006482.1 Arabidopsis thaliana At5g45300 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006398198.2 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | V4N342 | 0.0 | V4N342_EUTSA; Beta-amylase (Fragment) | ||||
| STRING | XP_006398198.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10001222m |
| Entrez Gene | 18016311 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



