 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10003256m | ||||||||
| Common Name | EUTSA_v10003256mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 279aa MW: 30970.8 Da PI: 9.7222 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.7 | 4.8e-32 | 109 | 162 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+av+ LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Thhalv10003256m 109 PRMRWTSTLHAHFVHAVQLLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 162
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.79E-15 | 106 | 162 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-28 | 107 | 162 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.9E-24 | 109 | 162 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.2E-7 | 110 | 161 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 279 aa Download sequence Send to blast |
MMMLESRSNM RGLKSVPDLS LQISLPNSHA LKPLHGGDRS STTSSDSGSS LSELSHENNF 60 FNKPLLSLGF DHHRRQQRHS HMFQPQIYGR DFKRSSSSMV CLKRSIRAPR MRWTSTLHAH 120 FVHAVQLLGG HERATPKSVL ELMNVKDLTL AHVKSHLQMY RTVKCTDKGS SGAGKVEKEA 180 DQRIEDNNNN NEEGTDTNSP NSLSAQKTER ASCSSTKGVS MSISTQADPH LGATRHTKDD 240 GEKKDINAHL NLEFTLGRPS WGIVDYAEPS RDLTLLKC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 1e-16 | 110 | 164 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-16 | 110 | 164 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-16 | 110 | 164 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-16 | 110 | 164 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-16 | 110 | 164 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates carpel integuments formation. Required for the specification of polarity in the ovule inner integument. Modulates the content of flavonols and proanthocyanidin in seeds. {ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:19054366, ECO:0000269|PubMed:20444210}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00107 | PBM | Transfer from AT5G42630 | Download |
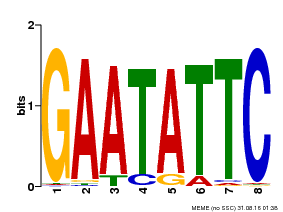
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10003256m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY048691 | 0.0 | AY048691.1 Arabidopsis thaliana GARP-like putative transcription factor KANADI4 (KAN4) mRNA, complete cds. | |||
| GenBank | BT026053 | 0.0 | BT026053.1 Arabidopsis thaliana At5g42630 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006403368.1 | 0.0 | probable transcription factor KAN4 | ||||
| Swissprot | Q9FJV5 | 1e-156 | KAN4_ARATH; Probable transcription factor KAN4 | ||||
| TrEMBL | V4NFC2 | 0.0 | V4NFC2_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006403368.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6572 | 28 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42630.1 | 1e-138 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10003256m |
| Entrez Gene | 18020634 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



