 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10005529m | ||||||||
| Common Name | EUTSA_v10005529mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 502aa MW: 55655.3 Da PI: 6.9687 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 32.6 | 2e-10 | 237 | 293 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + rk + g ++ +++Aa+a++ a++k++g
Thhalv10005529m 237 SIYRGVTRHRWTGRYEAHLWDnSCR-RegqARKGRQ-GGYDKEDKAARAYDLAALKYWG 293
57*******************4444.2344336655.7799999*************98 PP
| |||||||
| 2 | AP2 | 49.9 | 7.9e-16 | 336 | 387 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Thhalv10005529m 336 SIYRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 387
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.2E-8 | 237 | 293 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.63E-13 | 237 | 302 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 7.45E-18 | 237 | 303 | No hit | No description |
| PROSITE profile | PS51032 | 16.898 | 238 | 301 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 9.3E-20 | 238 | 307 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.2E-11 | 238 | 301 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.1E-6 | 239 | 250 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.49E-11 | 336 | 395 | No hit | No description |
| SuperFamily | SSF54171 | 9.15E-18 | 336 | 396 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 3.3E-10 | 336 | 387 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.9E-31 | 337 | 401 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.4E-18 | 337 | 395 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.111 | 337 | 395 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.1E-6 | 377 | 397 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 502 aa Download sequence Send to blast |
MANWLTFSLS PMEMLKSPDQ SHFVSYDASS APYLIDNLYV LKEEAQIETA SMADSTTLAT 60 FFDSQTHSPT QVPKLEDFFG DSSTSFVRYS DNQTERQNSS SLTRIYDPRH HTGEVTGYFA 120 DHHHDFKTIA CFQTNSGSEI VDDSASIGRA HLSKGGEFHV VDSSTTATAE LGFHGGCTNG 180 GALSLAVNNT DHPLSRNHGE RGKNSKKTIV SKKDTKAVES TDDSKKKIAE TFGQRTSIYR 240 GVTRHRWTGR YEAHLWDNSC RREGQARKGR QGGYDKEDKA ARAYDLAALK YWGSAATTNF 300 QIASYSKELE EMNHMTKQEF IASLRRTSSG FSRGASIYRG VTRHHQQGRW QARIGRVAGN 360 KDLYLGTFAT EEEAAEAYDI AAIKFRGINA VTNFEMNRYD VEAVMKSSFP VGGGGAAKRH 420 KLSLESPPPS SSSDRNLQQH LPPSSSSELD PNSIPCGIPF ESSVLHHHQD LFQHYPAVSD 480 STTVDAPMNQ AEFFLWPNHS Y* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00589 | DAP | Transfer from AT5G65510 | Download |
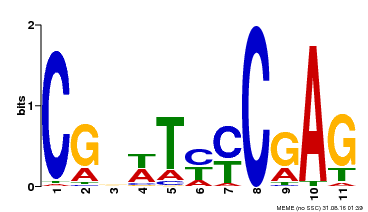
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10005529m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353404 | 0.0 | AK353404.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-30-G24. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024007164.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL7 | ||||
| Swissprot | Q6J9N8 | 0.0 | AIL7_ARATH; AP2-like ethylene-responsive transcription factor AIL7 | ||||
| TrEMBL | V4KMZ1 | 0.0 | V4KMZ1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006394021.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65510.1 | 0.0 | AINTEGUMENTA-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10005529m |
| Entrez Gene | 18011874 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



