 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10007000m | ||||||||
| Common Name | EUTSA_v10007000mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 668aa MW: 73870.5 Da PI: 5.9311 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 99 | 4e-31 | 64 | 148 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+++e+laL+++r++m++++r++ lk+plWe+vs+k++e g++rs+k+Ckek+en++k+yk++ke++++r++++ +++f+qlea
Thhalv10007000m 64 RWPREETLALLRIRSDMDSTFRDATLKAPLWEHVSRKLMELGYKRSAKKCKEKFENVQKYYKRTKETRGGRHDGK--AYKFFSQLEA 148
8*********************************************************************86555..5******985 PP
| |||||||
| 2 | trihelix | 105.1 | 5.1e-33 | 433 | 517 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+k e+laLi++r+ me r++++ k+ lWee+s+ m++ g++r++k+Ckekwen+nk+ykk+ke++kkr +++ +tcpyf++l+
Thhalv10007000m 433 RWPKAEILALINLRSGMEPRYQDNVPKGLLWEEISASMKRMGYNRNAKRCKEKWENINKYYKKVKESNKKR-PQDAKTCPYFHRLD 517
8*********************************************************************8.99999*******97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 6.946 | 57 | 121 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.11 | 61 | 123 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 3.4E-21 | 63 | 149 | No hit | No description |
| CDD | cd12203 | 1.14E-25 | 63 | 128 | No hit | No description |
| PROSITE profile | PS50090 | 6.876 | 426 | 490 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0069 | 430 | 492 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 4.3E-22 | 432 | 517 | No hit | No description |
| CDD | cd12203 | 3.88E-29 | 433 | 497 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0010090 | Biological Process | trichome morphogenesis | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0042631 | Biological Process | cellular response to water deprivation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:2000037 | Biological Process | regulation of stomatal complex patterning | ||||
| GO:2000038 | Biological Process | regulation of stomatal complex development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 668 aa Download sequence Send to blast |
MEQGGGGEVG GGGGGNEVVE EASPISSRPP ANLEELMRFS AAADDGGGLG GGGGGSSSSS 60 SGNRWPREET LALLRIRSDM DSTFRDATLK APLWEHVSRK LMELGYKRSA KKCKEKFENV 120 QKYYKRTKET RGGRHDGKAY KFFSQLEALN TTPPPLPPPP PPSSSLDVAP LSVANPIQPQ 180 SQFPVFPLPQ TQPQPSQPHL THTVSFTTPP PPLPPPPMGP TFPGVTFSSH SSSTASGMGS 240 DDDDDMDLDE AGPSSRKRKR GNRGGGEGGK MMELFEGLVR QVMEKQAAMQ RSFLDALEKR 300 EQERLQREEA WKRQEMSRLA REHEVMSQER AASASRDAAI ISLIQKITGH TIQLPPSLSS 360 SQPPQPPHQP PPPAAKRVEP PQTTTTQSQS QPIMAIPQQQ ILPPPHPPAP PPAPHQQQEM 420 IMSPEQSSPS SSRWPKAEIL ALINLRSGME PRYQDNVPKG LLWEEISASM KRMGYNRNAK 480 RCKEKWENIN KYYKKVKESN KKRPQDAKTC PYFHRLDLLY RNKVLGNGSG SSTSGLPQEQ 540 TSTVQKQSPV SKPPQEGVVN IHQPHGSASS EEEERIEESL QGTEKPEDIV MRELMQQQQQ 600 QQESMIGEYE KIEESHNYNN MEEDEEELDE EELDEDEKSA AYEIAFQSPA NRGGNGHTEP 660 PFLTMVQ* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 106 | 114 | KRSAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds specific DNA sequence such as GT3 box 5'-GGTAAA-3' in the SDD1 promoter. Negative regulator of water use efficiency (WUE) via the promotion of stomatal density and distribution by the transcription repression of SDD1. Regulates the expression of several cell cycle genes and endoreduplication, especially in trichomes where it prevents ploidy-dependent plant cell growth. {ECO:0000269|PubMed:19717615, ECO:0000269|PubMed:21169508}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00177 | DAP | Transfer from AT1G33240 | Download |
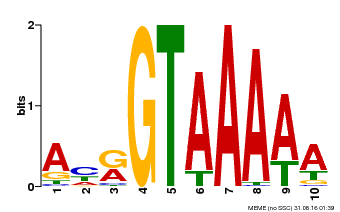
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10007000m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by water stress. {ECO:0000269|PubMed:21169508}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY140085 | 0.0 | AY140085.1 Arabidopsis thaliana DNA-binding factor, putative (At1g33240) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006415119.1 | 0.0 | trihelix transcription factor GTL1 isoform X1 | ||||
| Swissprot | Q9C882 | 0.0 | GTL1_ARATH; Trihelix transcription factor GTL1 | ||||
| TrEMBL | V4KTY6 | 0.0 | V4KTY6_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006415119.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6226 | 27 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G33240.1 | 8e-98 | GT-2-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10007000m |
| Entrez Gene | 18991735 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



