 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10007139m | ||||||||
| Common Name | EUTSA_v10007139mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 598aa MW: 66645.8 Da PI: 6.8577 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.9 | 7.6e-13 | 440 | 485 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L Av YI++L
Thhalv10007139m 440 NHVEAERQRREKLNQRFYALRSVVPNiS------KMDKASLLGDAVSYINEL 485
799***********************66......***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.2E-51 | 48 | 236 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.326 | 436 | 485 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.70E-14 | 439 | 490 | No hit | No description |
| SuperFamily | SSF47459 | 9.42E-19 | 439 | 506 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.0E-10 | 440 | 485 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.2E-18 | 440 | 499 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.8E-16 | 442 | 491 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 598 aa Download sequence Send to blast |
MNIGGVAWNE DDKAIVASLL GKRALDYLLS NSVPNANLLM TVGSDENLQN KLSDLVERPN 60 ASNFSWNYAI FWQISRSKAG DLVLCWGDGS CREPKEGEKS EIVRILSMGR EEETHQTMRK 120 RVLQKLHALF GGLEEDNCAL GLDRVTDTEM FLLASMYFSF PRGEGGPGKC FDSGKPVWLP 180 DVVNSGSDYC VRSFLAKSAG IQTIVLVPTD IGVVELGSTR SLPESQESML SIRSLFSSYL 240 PPPVRVVTPA ALPVVVADKN DDNRTVNSSK IFGKDLQNSG FLRHHQQQPQ QQQQQHRQFR 300 EKLTVRKMDD RVPKRLDTYP NNGNRYVFSN PSTNNNNNNS TLLSPTWVQP EMYTRANSVK 360 EVPNTEDFKF LPLQQSSQRL LPPAQMQIDF SGASSRAPEN NSDGEGGAEW ADVVGGDESG 420 NNKPRKRGRR PANGRAEALN HVEAERQRRE KLNQRFYALR SVVPNISKMD KASLLGDAVS 480 YINELHAKLK VMEAERERLG YSSNPPISLE PEINVQTSGE DVTVRVNCPL DSHPASRIFH 540 AFEEAKVEVI NSNMEFSQDT VLHAFVIRSE ELTKEKLISA FSREQSSSVQ SRTSSGR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 5e-26 | 434 | 496 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
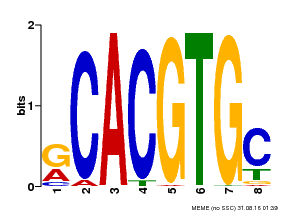
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10007139m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353331 | 0.0 | AK353331.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-24-P08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006418406.1 | 0.0 | transcription factor bHLH13 | ||||
| Swissprot | Q9LNJ5 | 0.0 | BH013_ARATH; Transcription factor bHLH13 | ||||
| TrEMBL | E4MXT4 | 0.0 | E4MXT4_EUTHA; mRNA, clone: RTFL01-24-P08 | ||||
| TrEMBL | V4KZG7 | 0.0 | V4KZG7_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006418406.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10007139m |
| Entrez Gene | 18994660 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



