 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10007196m | ||||||||
| Common Name | EUTSA_v10007196mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 578aa MW: 64289.6 Da PI: 4.6609 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 157.2 | 7.1e-49 | 18 | 144 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevl 93
+pGfrFhPtdeelvv+yLk+kv g+kl++ ++i vd+ykv+P++Lp + +k+++++w++F++r++ky++ r++r t++gyWkatgkd+ ++
Thhalv10007196m 18 APGFRFHPTDEELVVYYLKRKVCGRKLRI-NAIGVVDVYKVDPSELPglSVLKSGDRQWFYFTPRSRKYPNAARSSRGTATGYWKATGKDRVIV 110
59***************************.89***************644888899************************************** PP
NAM 94 skkgelvglkktLvfykgrapkgektdWvmheyrl 128
+++ vglkktLvfy+grap+ge+tdWvmhey++
Thhalv10007196m 111 Y-NSRPVGLKKTLVFYRGRAPNGERTDWVMHEYTM 144
9.999****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 52.725 | 17 | 167 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 7.85E-56 | 17 | 167 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.0E-25 | 19 | 143 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900057 | Biological Process | positive regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 578 aa Download sequence Send to blast |
MANSSSPDSC LRGGKFNAPG FRFHPTDEEL VVYYLKRKVC GRKLRINAIG VVDVYKVDPS 60 ELPGLSVLKS GDRQWFYFTP RSRKYPNAAR SSRGTATGYW KATGKDRVIV YNSRPVGLKK 120 TLVFYRGRAP NGERTDWVMH EYTMDEEELG RCKNVKDYYA LYKLYKKSGA GPKNGEEYGA 180 PFQEEEWVDD DDSEDADNVV AVPDDHVVRY ENNCRVDDGM FCNPVSVQLD DLEKLLNEIP 240 DAPGVAPREM SDLSGVLQGN SEGEIQSTLL NNFPGEFLAP REVGKFSPNC QPYNRPLSFQ 300 PHLRSENSAF NASGMAPLVF EKEDYIEMDD LLIPELRAST TEESSQFLNH GELGNVNEFD 360 QLFHDISMSL DAEPVFQGTS TDLSSLSDHA SNTSDQRQQF LYQEVQDQSP ENQLNNFIDD 420 PSTSLNQFTD DMWFHDAQAD LFDQPQSSSL AFASPSPGVI PGSTNPTMNV NAQDQEGQNG 480 VGKTSQFSSA LWAFMESIPS TPASACEGPL NRTFVRMSSF SRIRFNGTSV TTSKVTKAKK 540 RMGNRGFLLL SIMGALCAIF WVFIIATVGV LGRPILS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 9e-44 | 19 | 144 | 17 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (By similarity). Transcriptional activator that promotes leaf senescence by up-regulating senescence-associated genes in response to developmental and stress-induced senescence signals. Functions in salt and oxidative stress-responsive signaling pathways. Binds to the promoter of NAC029/NAP and NAC059/ORS1 genes (PubMed:23926065). {ECO:0000250|UniProtKB:Q949N0, ECO:0000269|PubMed:23926065}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00178 | DAP | Transfer from AT1G34180 | Download |
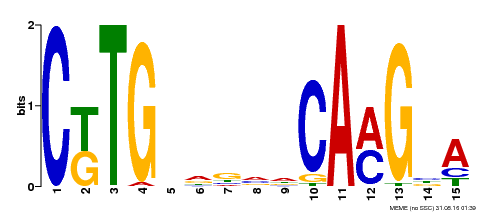
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10007196m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, salt, drought stress and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006415017.1 | 0.0 | NAC domain-containing protein 16 | ||||
| Refseq | XP_024008436.1 | 0.0 | NAC domain-containing protein 16 | ||||
| Swissprot | A4FVP6 | 0.0 | NAC16_ARATH; NAC domain-containing protein 16 | ||||
| TrEMBL | V4K6Y9 | 0.0 | V4K6Y9_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006415017.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2812 | 26 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34180.1 | 0.0 | NAC domain containing protein 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10007196m |
| Entrez Gene | 18991637 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



