 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10008930m | ||||||||
| Common Name | EUTSA_v10008930mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 170aa MW: 19852.5 Da PI: 9.0476 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 77.4 | 1.9e-24 | 3 | 53 | 2 | 58 |
ZF-HD_dimer 2 ekvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreve 58
++++Y eC++NhAa+lG++avDGC+E+ + a++ CaACgCHR++HRr +
Thhalv10008930m 3 SSCVYSECMRNHAAKLGSYAVDGCREYSQP------ATGDLCAACGCHRSYHRRIDV 53
5789***********************998......57789*************866 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 4.0E-20 | 4 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 2.3E-17 | 6 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 1.3E-15 | 7 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 17.49 | 7 | 51 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene3D | G3DSA:1.10.10.60 | 8.2E-17 | 93 | 166 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.34E-7 | 99 | 165 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 6.9E-30 | 107 | 166 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 170 aa Download sequence Send to blast |
MQSSCVYSEC MRNHAAKLGS YAVDGCREYS QPATGDLCAA CGCHRSYHRR IDVKSSGQIT 60 RAPFPFTSLR RVKQLARLKW RATEEREEEE EEEDTEETST EEMMTVKRRR KSKFTAEQKE 120 AMRDYAVKLG WTLKDKRAIR EEIRVFCEGI GVSRYLFKTW VNNNKKFYH* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 107 | 112 | RRRKSK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00143 | DAP | Transfer from AT1G14687 | Download |
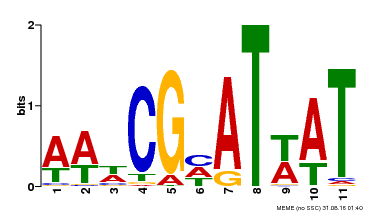
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10008930m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006416985.1 | 1e-124 | zinc-finger homeodomain protein 14 | ||||
| Swissprot | Q9LQW3 | 1e-92 | ZHD14_ARATH; Zinc-finger homeodomain protein 14 | ||||
| TrEMBL | V4L6F0 | 1e-123 | V4L6F0_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006416985.1 | 1e-124 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13728 | 21 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14687.1 | 9e-93 | homeobox protein 32 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10008930m |
| Entrez Gene | 18993398 |



