 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10010066m | ||||||||
| Common Name | EUTSA_v10010066mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1379aa MW: 154581 Da PI: 7.6615 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.4 | 0.00096 | 1261 | 1286 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sF++ +L H r+ +
Thhalv10010066m 1261 YQCDmeGCTMSFTSEKQLALHKRNiC 1286
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 13.1 | 0.00028 | 1286 | 1308 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Thhalv10010066m 1286 CPvkGCGKNFFSHKYLVQHRRVH 1308
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 11 | 0.0013 | 1344 | 1370 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Thhalv10010066m 1344 YVCAepSCGQTFRFVSDFSRHKRKtgH 1370
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.6E-16 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.077 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 5.7E-14 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 3.5E-52 | 201 | 370 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 34.061 | 204 | 370 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 3.84E-27 | 219 | 388 | No hit | No description |
| Pfam | PF02373 | 2.8E-37 | 234 | 353 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 2.5E-4 | 1260 | 1283 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 8.5 | 1261 | 1283 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.016 | 1284 | 1308 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.965 | 1284 | 1313 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.0E-6 | 1285 | 1307 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1286 | 1308 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.55E-10 | 1300 | 1342 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.5E-10 | 1308 | 1335 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 1314 | 1338 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.928 | 1314 | 1343 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1316 | 1338 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 8.38E-8 | 1332 | 1366 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-8 | 1336 | 1367 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.5 | 1344 | 1370 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.281 | 1344 | 1375 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1346 | 1370 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1379 aa Download sequence Send to blast |
MAVSEQSPDV FPWLKSLPVA PEFRPTLAEF QDPIAYIFKI EEEASKYGIC KILPPVPPSS 60 KKTAINNLNR SLAARAAARV RDGADDGAWD YDGAPTFATR QQQIGFCPRK QRPVQRPVWQ 120 SGEHYSFGEF EFKAKNFEKN YLRKCGKRSQ LSALEIETLY WRATVDKPFS VEYANDMPGS 180 AFIPLSLAAA RRRESGGDGG TVGETAWNMR AMARAEGSLL KFMKEEIPGV TSPMVYIAMM 240 FSWFAWHVED HDLHSLNYLH MGAGKTWYGV PKDAAVAFEE VVRVHGYGGE LNPLVTFSTL 300 GEKTTVMSPE VFVRAGIPCC RLVQNPGEFV VTFPGAYHSG FSHGFNFGEA SNIATPQWLR 360 MAKDAAIRRA AINYPPMVSH LQLLYDFALA LGSRVPTSIN TKPRSSRLKD KKRSEGEKLT 420 KELFVQNIIH NNELLCSLGK GSPVALLPQS SSDVSVCSDL RIGSHLRTNQ EKPIQLKSED 480 LSSDSIMPGL NNGFKDAVSV KEKFTSLCER NRNHLASREN ETQGTSTDAE RKKNDRAVGL 540 SDQRLFSCVT CGILSFDCVA IVQPKEAAAR YLMSADCSFF NDWTVASGPA NLGQVAGSLH 600 PQSTGKHDVD YFYNVPVQTT DHSMKTGDQR TSSSSLTKEN KDDGALGLLA SAYGDSSDSE 660 EEDHKGSDIP ISEGITRQYD QEGACVSEAT SFDTDRNEEA RDGPSSDINS QRLTSGKGKE 720 IDVLHATSSC STLSCTSLQE IALPFIPRSD DDSSRLHVFC LEHAAEVEQQ LRPIGGIRIM 780 LLCHPEYPRI EAEAKIVSEE LGVNHEWNDT EFRNVSRVDE ETIQAALDNV EAKAGNSDWS 840 VKLGINLSYS AILSRSPLYS KQMPYNSVIY NAFGRGSPAT SSPTKPQVSG KRSSRQRKYV 900 VGKWCGKVWM SHQVHPFLLQ QDLEGDESER SCHLRGVLED DVIGKRLSPC NASRDATTMF 960 GRKYCRKRKM RAKALPRKKL TSFKREDVVS DDTSEDHSYK QQWRASGNEE ESYFETGNSV 1020 SGDSSNQMSD EQQEFVRHQG DKDFESDDEV SDRSLGEEYA VRECAASESS MENGSQLYRE 1080 NQSMYDHDDD DIYRHPRGLP RSKRTKFFRN PVSYDSEENG LYQQRGRVST SNAQISRMGG 1140 AYDSADNSLE EQEFCSAGKR QTRSTAKRKV KTEIVQSLRD TKRRALRQSG PRKKNQEVDI 1200 YMEGPSTRLR VRNLNPSIGS SETKPKKTGK KRNSNSFSRV ASEEELEEDE EENEQEECAP 1260 YQCDMEGCTM SFTSEKQLAL HKRNICPVKG CGKNFFSHKY LVQHRRVHSD DRPLKCPWKG 1320 CKMTFKWAWS RTEHIRVHTG ARPYVCAEPS CGQTFRFVSD FSRHKRKTGH SVKKTKKR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 2e-77 | 1258 | 1378 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 2e-77 | 1258 | 1378 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 2e-77 | 1258 | 1378 | 20 | 140 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 966 | 978 | KRKMRAKALPRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
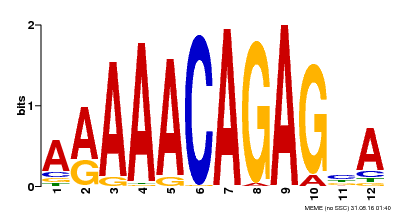
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10010066m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006404261.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | V4LPK0 | 0.0 | V4LPK0_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006404261.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10010066m |
| Entrez Gene | 18020751 |



