 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10011463m | ||||||||
| Common Name | EUTSA_v10011463mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 460aa MW: 51016.2 Da PI: 8.5981 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.3 | 4.8e-12 | 396 | 441 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR +i +++ +L++l+P+ ++ + a++L +Av+YIk+Lq
Thhalv10011463m 396 SIAERVRRTKISERMRKLQDLVPNM----DTQTNTADMLDLAVQYIKDLQ 441
689*********************7....79******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.072 | 390 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.83E-16 | 393 | 456 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.77E-11 | 394 | 445 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.1E-15 | 395 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.3E-9 | 396 | 441 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.3E-14 | 396 | 446 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 460 aa Download sequence Send to blast |
MFVFVPSKKN DIKLPSRSLS PLFFSFFFPF LVFFISHILI YNLRDLSPGK SPGESAKLPV 60 LSNPTEKITR EEKNTKATKM ESEFQQHHFH LHDHQHQRPR NSGLIRYQSA PSSYFSSFGD 120 RESIEEFLDR PTSPETERIL SGFLQTTDTS NNVDSFLHHT FNSDGSEKKP PEVKMEEVDI 180 PATAMEVVVS GGGDDVEVSV APDSIGYDSV VRNLGQNKRP RERDDRTPAN NLARHNSSPA 240 GLFSSVDVET AYAAVMKSMG GFGGGNLMNT SNTEASSFTP RSKLPPSASR QMSPISEIDV 300 KPGFSSRLPP RTLSGGFNRS FGNEGSASSK LSAIARTQSG GLDQYKTKDE DSASRRPPLA 360 HHMSLPKSLS DIEQLLSDSI PCKIRAKRGC ATHPRSIAER VRRTKISERM RKLQDLVPNM 420 DTQTNTADML DLAVQYIKDL QEQVKELEES RARCRCSSA* |
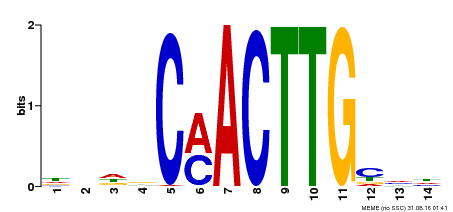
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00195 | DAP | Transfer from AT1G51140 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10011463m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY034941 | 0.0 | AY034941.1 Arabidopsis thaliana unknown protein (At1g51140) mRNA, complete cds. | |||
| GenBank | AY063120 | 0.0 | AY063120.1 Arabidopsis thaliana unknown protein (At1g51140) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024006340.1 | 0.0 | transcription factor bHLH122 | ||||
| Swissprot | Q9C690 | 0.0 | BH122_ARATH; Transcription factor bHLH122 | ||||
| TrEMBL | V4KKA9 | 0.0 | V4KKA9_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006393076.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10547 | 26 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51140.1 | 0.0 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10011463m |
| Entrez Gene | 18010834 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



