 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10011650m | ||||||||
| Common Name | EUTSA_v10011650mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 327aa MW: 35584.9 Da PI: 9.3235 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 27.8 | 6e-09 | 7 | 54 | 3 | 45 |
SS-HHHHHHHHHHHHHTTTT......-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg......tWktIartmgkgRtlkqcksrwq 45
W+ eE++ + +a++++ + +W++ a++++ +++ +++k ++q
Thhalv10011650m 7 TWSREEEKAFENAIAMHCVEeeltddQWTKMASMVP-TKSSEEVKKHYQ 54
6****************999****************.***********9 PP
| |||||||
| 2 | Myb_DNA-binding | 38.7 | 2.2e-12 | 141 | 185 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + ++G+g+W++I+r + Rt+ q+ s+ qky
Thhalv10011650m 141 PWTEEEHRLFLLGLDKFGKGDWRSISRNFVISRTPTQVASHAQKY 185
7*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 8.995 | 3 | 61 | IPR017884 | SANT domain |
| SMART | SM00717 | 9.1E-8 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.7E-5 | 6 | 58 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.1E-7 | 7 | 54 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.09E-8 | 7 | 57 | No hit | No description |
| SuperFamily | SSF46689 | 7.18E-9 | 8 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.873 | 134 | 190 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.61E-17 | 136 | 191 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.7E-17 | 138 | 188 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.3E-10 | 138 | 188 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.3E-11 | 139 | 184 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.15E-9 | 141 | 186 | No hit | No description |
| Pfam | PF00249 | 8.3E-11 | 141 | 185 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
METVAVTWSR EEEKAFENAI AMHCVEEELT DDQWTKMASM VPTKSSEEVK KHYQILLEDV 60 KAIENGHVPL PRYHRKGIAI DEAAAAATSP ANRDGHSSGG SGSSEKKPNH GASGTSSSNG 120 GGGGGGRSGS RAEQERRKGI PWTEEEHRLF LLGLDKFGKG DWRSISRNFV ISRTPTQVAS 180 HAQKYFIRLN SMNRDRRRSS IHDITTVNNQ APAVTGQQQQ QQQQQQVVKH RPAQPQPQPQ 240 PQPQPPHHPG ATMAGLGMYG GAPMGQPIIA PPDHMGSAVG TPVMLPPPMV PHHHHHHHHL 300 GVAPYPVPAY PVPPIPQQHP APSTMH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 8e-14 | 8 | 77 | 11 | 77 | RADIALIS |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00191 | DAP | Transfer from AT1G49010 | Download |
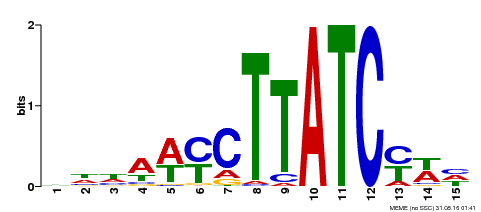
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10011650m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006393319.1 | 0.0 | transcription factor MYBS1 | ||||
| TrEMBL | V4JYM2 | 0.0 | V4JYM2_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006393319.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10304 | 25 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49010.1 | 1e-123 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10011650m |
| Entrez Gene | 18011444 |



