 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10013367m | ||||||||
| Common Name | EUTSA_v10013367mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 493aa MW: 53935.1 Da PI: 7.2525 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.2 | 7.6e-17 | 164 | 213 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Thhalv10013367m 164 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAVKFRG 213
78*******************......55***********************9997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 2.42E-15 | 164 | 221 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 2.15E-11 | 164 | 220 | No hit | No description |
| Pfam | PF00847 | 1.2E-8 | 164 | 213 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.3E-30 | 165 | 227 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-16 | 165 | 221 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.359 | 165 | 221 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 493 aa Download sequence Send to blast |
MLDLNLDVDS AESTQNGRDS AAVKRVSGAI LNQMDESVTS NSSVVNAEAS SCIDGEEELC 60 STRAVKFQFE ILKGGGGKGE EEEEEEVEER SAVMTKEFFP VAKGDGEGMY FLDSSAQSSR 120 CPVDISFQRG NLGGDFPGGD SAPVMQPPSQ PVKKSRRGPR SKSSQYRGVT FYRRTGRWES 180 HIWDCGKQVY LGGFDTAHAA ARAYDRAAVK FRGLEADINF IISDYEEDLK QMANLSKEEV 240 VQVLRRQSSG FSRNNSRYQG VALQKIGNWG AQMEQFHGNM ACDKAAIKWN GREAASLIEP 300 HASRMTPEAA NVKLDLNLGI SLSLGDGPKQ KDRGLRLHHA PNSIVCGRNS KMENHMAATT 360 CDTSFNFLKR GSEHLINRHV LPSAFFSPME RTPEKGFMPR SHQSFPARTW QVQDQSSSGT 420 ATTATASPLF SNAASSGFSL SATRPPSSST ATLHPSHPFL NLNMPGLYVI HPSDYASQQH 480 HPHLMNRSQP PP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00616 | PBM | Transfer from AT5G60120 | Download |
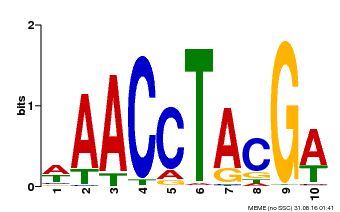
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10013367m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352946 | 0.0 | AK352946.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-19-B14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006400879.1 | 0.0 | AP2-like ethylene-responsive transcription factor TOE2 | ||||
| Swissprot | Q9LVG2 | 0.0 | TOE2_ARATH; AP2-like ethylene-responsive transcription factor TOE2 | ||||
| TrEMBL | E4MWP9 | 0.0 | E4MWP9_EUTHA; mRNA, clone: RTFL01-19-B14 | ||||
| TrEMBL | V4LIS4 | 0.0 | V4LIS4_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006400879.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM12836 | 17 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G60120.1 | 0.0 | target of early activation tagged (EAT) 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10013367m |
| Entrez Gene | 18018477 |



