 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10019126m | ||||||||
| Common Name | EUTSA_v10019126mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 215aa MW: 23473.8 Da PI: 10.2733 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 139.4 | 4e-43 | 48 | 190 | 4 | 163 |
YABBY 4 fssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnve.keesastsv 96
s++e++ yv+C+ Cntilav +P + ++ +vtv+CGhC +l ++ + + + h + +l+++ ++ +e k+ s+s+s
Thhalv10019126m 48 SSQAEHLYYVRCSICNTILAVGIPLKRMLDTVTVKCGHCGNLSFLTTSP-----PLQGH--------VSLTLQMQ--SFGGSHEyKKGSSSSSS 126
5789************************************975533332.....22232........33333333..33333220333333333 PP
YABBY 97 sseklsenedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahf 163
ss +++++ +pr+p v++PPek+qr Psaynrf+++eiqrik++nP+i hreafsaaaknWa +
Thhalv10019126m 127 SS---TSSDQPPSPRPPFVVKPPEKKQRLPSAYNRFMRDEIQRIKSANPEIPHREAFSAAAKNWAKY 190
33...3457788999999***********************************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 6.8E-50 | 50 | 190 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.7E-8 | 134 | 189 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 9.5E-5 | 144 | 189 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010254 | Biological Process | nectary development | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048479 | Biological Process | style development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 215 aa Download sequence Send to blast |
MWASQGLLAS NLNELHYSNS LSLNKRVLIH SIKIMNLEEK PTMASRASSQ AEHLYYVRCS 60 ICNTILAVGI PLKRMLDTVT VKCGHCGNLS FLTTSPPLQG HVSLTLQMQS FGGSHEYKKG 120 SSSSSSSSTS SDQPPSPRPP FVVKPPEKKQ RLPSAYNRFM RDEIQRIKSA NPEIPHREAF 180 SAAAKNWAKY IPNSPTSITS GGNNINGLGF GEKK* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for the initiation of nectary development. Also involved in suppressing early radial growth of the gynoecium, in promoting its later elongation and in fusion of its carpels by regulating both cell division and expansion. Establishes the polar differentiation in the carpels by specifying abaxial cell fate in the ovary wall. Regulates both cell division and expansion. {ECO:0000269|PubMed:10225997, ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:10535738, ECO:0000269|PubMed:11714690, ECO:0000269|PubMed:15598802, ECO:0000269|Ref.10}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00220 | DAP | Transfer from AT1G69180 | Download |
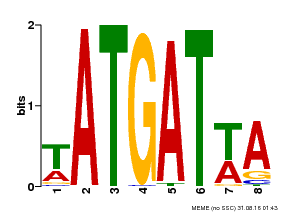
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10019126m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by SPT and by A class genes AP2 and LUG in the outer whorl. In the third whorl, B class genes AP3 and PI, and the C class gene AG act redundantly with each other and in combination with SEP1, SEP2, SEP3, SHP1 and SHP2 to activate CRC in nectaries and carpels. LFY enhances its expression. {ECO:0000269|PubMed:10225998, ECO:0000269|PubMed:15598802}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ119055 | 0.0 | DQ119055.1 Brassica juncea transcription factor CRC mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024007404.1 | 1e-132 | protein CRABS CLAW | ||||
| Swissprot | Q8L925 | 1e-106 | CRC_ARATH; Protein CRABS CLAW | ||||
| TrEMBL | V4KB50 | 1e-157 | V4KB50_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006391053.1 | 1e-158 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9725 | 28 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69180.1 | 1e-108 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10019126m |
| Entrez Gene | 18008251 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



