 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10024342m | ||||||||
| Common Name | EUTSA_v10024342mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 918aa MW: 104029 Da PI: 7.334 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162.9 | 5.7e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
+e +rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++nptf
Thhalv10024342m 30 EEaYSRWLRPNEIHALLCNHKYFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGDDNPTFV 123
55589***************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcywlL++++e+ivlvhy+e++
Thhalv10024342m 124 RRCYWLLDKSQEHIVLVHYRETH 146
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 77.304 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.9E-75 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.8E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 6.02E-13 | 369 | 455 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.43E-12 | 555 | 665 | No hit | No description |
| Pfam | PF12796 | 5.8E-7 | 556 | 635 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.4E-15 | 556 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.06E-14 | 564 | 670 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.681 | 573 | 638 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 7.6E-6 | 606 | 635 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 606 | 638 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.47E-5 | 720 | 821 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.181 | 755 | 781 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 770 | 792 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 4.7E-4 | 793 | 815 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.651 | 794 | 818 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 6.4E-4 | 796 | 815 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.8 | 869 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.37 | 870 | 899 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.22 | 871 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 918 aa Download sequence Send to blast |
MTGDDSGKLL GSEIHGFHTL QDLDIQTMLE EAYSRWLRPN EIHALLCNHK YFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEVQA APATPGNSYS SSITDHLSPK LVSEDINSGV 180 RNTCNTGYEV RSNSMVARNH EIRLHEINTL DWDELLVPAE ISNQCPPTED DVLYFTEQLQ 240 TAPRGSAKQG NHLAGYNGST DVPSYLGLGD PVYQNKNPCV AGEFSSQHSH CVADPNPSAT 300 VGDQAGDALL NNGYGSQESF GRWVNNFISD SPGSVEDPSL DGVFTPGQDS STTPASFDSH 360 SNIPEQVFNI TDVSPAWAYS TEKTKILVTG YFHESFQHFG RSNLFCICGE LRVTAEFLQM 420 GVYRCFLPPQ SPGVVNLYLS ADGNNPISQL FSFEHRSVPV IEKAIPQNDQ LYKWEEFEFQ 480 VRLAHLLFTS SNKISVFSSK ISPDNLLEAK KLASRTSHLL NSWAYLIKSI QANEVPFDQA 540 RDHLFELTLK NRLKEWLLEK VIENRNTKEY DSKGLGVIHL CAVLGYTWSI LLFSWASISL 600 DFRDKNGWTA LHWAAFYGRE KMVAALLSAG ARPNLVTDPT KEYLGGCTAA DLAQQKGYDG 660 LAAFLAEKCL VAQFSDMKIA GNITGNLEGI KAETSTNPGN ANEEEQSLKD TLAAYRTAAE 720 AAARIQGAFR EHELKVRSSA VRFASKEEEA KNIIAAMKIQ HAFRNYETRR KIAAAARIQY 780 RFQTWKMRRE FLNMRNKAIR IQAAFRGFQV RRQYQKITWS VGVLEKAILR WRLKRKGFRG 840 LQVNQPEEKE GSDTVEDFYK TSQKQAEERL ERSVVKVQAM FRSKKAQQDY RRMKLAHEEA 900 QREYDGMQEL DQMDMES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
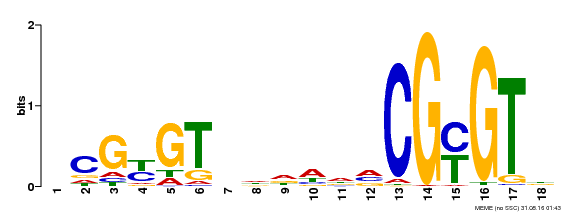
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10024342m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006414383.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | V4P888 | 0.0 | V4P888_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006414383.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10024342m |
| Entrez Gene | 18031060 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



