 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10024493m | ||||||||
| Common Name | EUTSA_v10024328mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 759aa MW: 83653.3 Da PI: 6.2344 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 92.3 | 3.7e-29 | 387 | 437 | 2 | 52 |
RWP-RK 2 ekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
ek+isl++l++yF +++kdAAk+Lgvc+T++KriCRq+GI+RWP+Rkik++
Thhalv10024493m 387 EKTISLDVLQQYFTGSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKV 437
799**********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.757 | 376 | 457 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 5.5E-26 | 389 | 437 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 2.49E-23 | 659 | 747 | No hit | No description |
| Gene3D | G3DSA:3.10.20.240 | 1.3E-27 | 662 | 743 | No hit | No description |
| Pfam | PF00564 | 1.4E-20 | 662 | 743 | IPR000270 | PB1 domain |
| SMART | SM00666 | 1.6E-28 | 662 | 744 | IPR000270 | PB1 domain |
| PROSITE profile | PS51745 | 27.83 | 662 | 744 | IPR000270 | PB1 domain |
| CDD | cd06407 | 3.15E-34 | 663 | 743 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 759 aa Download sequence Send to blast |
MISLTYMFSV DSESDIELGL PGRVFRQKLP EWTPNVQYYS SKEFSRLDHA LHYNVRGTLA 60 LPVFNLSGES CIGVVELIMT SEKIHYAPEV DRVCKALEAV NLKSSEILDH QTTQICNESR 120 QNALAEILEV LTVVCETHNL PLAQTWVPCQ HGSVLANGGG LKKNCTSFDG SCMGQVCMST 180 TDMACYVVDA HVWGFRDACL EHHLQKGQGV AGRAFLNGGS CFCRDITKFC KTQYPLVHYA 240 LMFKLTTCFA ISLQSSYTGD DSYILEFFLP SSITDDQEQD ALLGSLLVTM KEHFQSLRVA 300 SGVDFGEDDD KLSFEIIRAL PDKKIHSKIE SIRVPFSGFK SNATETLLIP QHAVQSSDPV 360 NEKSNVATVN GVVKEKKKTE KKRGKTEKTI SLDVLQQYFT GSLKDAAKSL GVCPTTMKRI 420 CRQHGISRWP SRKIKKVNRS ITKLKRVIES VQGTDGGLDL TSMAVSSIPW THGHQTAAQP 480 LNSPSGSKPP ELPNTDNSPN HWSSDHSPHE PNGSPELPNN GHKRSRTADE SAGTPTSHGS 540 CDGNQLDETK VLNQDPLFTV GGSPGLFFPP YSRDHDVSAA SFSLPNRLLG TIDHFRGMPI 600 EDAGSSKDLR NLCSTAAFED KFPDSNWMTN DNNSNNNNIY APPKEEAIVN ITREPSGSEM 660 RTVTIKASYK EDIIRFRISS GSGIMELKDE VAKRLKLDAG TFDIKYLDDD NEWVLIACDA 720 DLQECLEIPR SSRTNIVRLL VHDATTNLGS SCESTGEL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in regulation of nitrate assimilation and in transduction of the nitrate signal. {ECO:0000269|PubMed:18826430}. | |||||
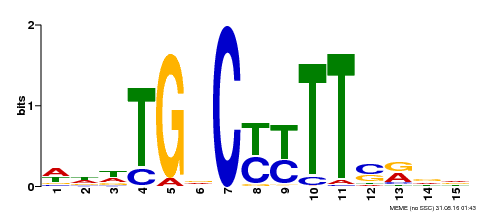
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10024493m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by the N source or by nitrate. {ECO:0000269|PubMed:18826430}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228452 | 0.0 | AK228452.1 Arabidopsis thaliana mRNA for hypothetical protein, complete cds, clone: RAFL15-03-P13. | |||
| GenBank | BT005776 | 0.0 | BT005776.1 Arabidopsis thaliana clone RAFL15-03-P13 (R20174) unknown protein (At4g24020) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006413484.1 | 0.0 | protein NLP7 | ||||
| Refseq | XP_024005488.1 | 0.0 | protein NLP7 | ||||
| Swissprot | Q84TH9 | 0.0 | NLP7_ARATH; Protein NLP7 | ||||
| TrEMBL | V4MPG8 | 0.0 | V4MPG8_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006413484.1 | 0.0 | (Eutrema salsugineum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10024493m |
| Entrez Gene | 18030707 |



