 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10024893m | ||||||||
| Common Name | EUTSA_v10024893mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 531aa MW: 60407.4 Da PI: 6.4607 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 39.8 | 7.8e-13 | 22 | 89 | 16 | 95 |
EE--HHH.HTT---..--SEEEEEE.TTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEE CS
B3 16 lvlpkkfaeehggkkeesktltled.esgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvk 95
l+l +f++++ + + l++ s r Wevkl ++++ l++GW+eF+ ++ ++gD++vF++d++++f+ v+
Thhalv10024893m 22 LTLDDEFLRHYTK-------VLLRSdASDRIWEVKL--NGNR----LAGGWEEFAAVHRFRDGDVLVFRHDKDENFHVSVG 89
6777777777753.......445443999*******..9998....***********************999898854444 PP
| |||||||
| 2 | B3 | 63.3 | 3.8e-20 | 156 | 252 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefel 92
f+++ +++++l ++rl l+++f+ + + +++ ++ l +e+gr+W++ l+ + + g +++++GW +F+++n+L++gD + Fkl++ +++++
Thhalv10024893m 156 FITAHVTPYSLIKDRLDLSRNFTVASfdE--DNKPCEIDLANEKGRKWTLLLRRNISTGVFYIRRGWTNFCSSNELRQGDLCKFKLSE-NGERP 246
7888999***************9888444..456779***************88888888*************************987.66668 PP
EEEEE- CS
B3 93 vvkvfr 98
v+ ++
Thhalv10024893m 247 VLWLCP 252
887775 PP
| |||||||
| 3 | B3 | 68.9 | 7e-22 | 295 | 392 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTE..EEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgr..yvltkGWkeFvkangLkegDfvvFkldgrsefel 92
f+k+ ++++ +ksg+l+++ f++e g+++++ ++tl +++gr+W+ +l +++k g+ ++l+kGWke++kang+k++D++v++l++ +++++
Thhalv10024893m 295 FLKIKLTPNRFKSGQLYISSVFVNESGINNAG--EITLLNKDGRKWSSYLHMTGKCGTewFYLRKGWKEMCKANGVKVNDSFVLELIW-EDANP 385
899999********************999766..8***************9977776678****************************.89999 PP
EEEEE-S CS
B3 93 vvkvfrk 99
++k+++k
Thhalv10024893m 386 TFKFYSK 392
*999887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01019 | 1.7E-12 | 10 | 92 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 7.19E-10 | 22 | 88 | No hit | No description |
| SuperFamily | SSF101936 | 5.49E-11 | 27 | 88 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 9.2E-12 | 33 | 88 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 10.771 | 34 | 92 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.6E-7 | 35 | 90 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 8.7E-18 | 150 | 253 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.75E-18 | 153 | 254 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 9.98E-18 | 154 | 252 | No hit | No description |
| SMART | SM01019 | 2.3E-24 | 156 | 254 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 15.185 | 156 | 253 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 6.5E-18 | 156 | 252 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.8E-16 | 294 | 387 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 8.83E-17 | 294 | 392 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 3.3E-26 | 295 | 393 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.38 | 295 | 393 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.2E-23 | 295 | 392 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 6.23E-16 | 295 | 391 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 531 aa Download sequence Send to blast |
MADPPNISLF HQRFLTGDNP LLTLDDEFLR HYTKVLLRSD ASDRIWEVKL NGNRLAGGWE 60 EFAAVHRFRD GDVLVFRHDK DENFHVSVGS RSDSCDMQHA SPSIVDTDDT EIDDDYVESA 120 DHHDDGEDED EDDAGDIVVS KNKKPEAESS SGDPCFITAH VTPYSLIKDR LDLSRNFTVA 180 SFDEDNKPCE IDLANEKGRK WTLLLRRNIS TGVFYIRRGW TNFCSSNELR QGDLCKFKLS 240 ENGERPVLWL CPQEYGNGHE EQECPRVSAE GGCSKEKSML DEIPKGKEKN TPSTFLKIKL 300 TPNRFKSGQL YISSVFVNES GINNAGEITL LNKDGRKWSS YLHMTGKCGT EWFYLRKGWK 360 EMCKANGVKV NDSFVLELIW EDANPTFKFY SKEENHENKG KGNRRTRKRR ACGTDLQGSS 420 EEKSRRVERE GRRVSRRAEH DEKEKRGNTQ VSNRPTTNSS KLQRKQAKPC SVSDQVANVK 480 RGILDTLKTV SQFRAELETR EKKLEAALLE IEALGERILG ISQILNNNLV * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May play a role in flower development. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00602 | PBM | Transfer from AT4G31610 | Download |
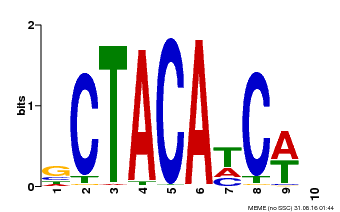
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10024893m |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006412578.1 | 0.0 | B3 domain-containing protein REM1 | ||||
| Swissprot | Q84J39 | 0.0 | REM1_ARATH; B3 domain-containing protein REM1 | ||||
| TrEMBL | V4P3P1 | 0.0 | V4P3P1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006412578.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9950 | 17 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31610.1 | 1e-165 | B3 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10024893m |
| Entrez Gene | 18030226 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



