 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Thhalv10027890m | ||||||||
| Common Name | EUTSA_v10027890mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Eutremeae; Eutrema
|
||||||||
| Family | GeBP | ||||||||
| Protein Properties | Length: 261aa MW: 29577.3 Da PI: 9.6453 | ||||||||
| Description | GeBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF573 | 95.3 | 6.2e-30 | 52 | 143 | 3 | 98 |
DUF573 3 fqrlwseeDeivlLqGlidfkaktgkspsddidafyefvkksisfkvsksqlveKirrLKkKfkkkvkkaksgkepsfskehdqkifelskkiW 96
++r+w+eeDe+++L+Gl+d+ +k+g++p +d+dafy+f+ +si k+sk+ql++Kir+LKkKf +++k+++g+++ d+++ ls++iW
Thhalv10027890m 52 SNRIWNEEDELTVLKGLVDYLDKEGVQPNSDWDAFYHFLGDSIAEKFSKQQLLSKIRKLKKKFLLHMEKTSQGDDMLC----DSEAIRLSRMIW 141
78*******************************************************************999999765....6788******** PP
DUF573 97 gs 98
g+
Thhalv10027890m 142 GQ 143
96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04504 | 5.2E-22 | 53 | 142 | IPR007592 | Protein of unknown function DUF573 |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 261 aa Download sequence Send to blast |
MAKPKQLDFS DDNSPSTRKS PRKRASNPSP ASETTKKKKK KKKTKKSAPP VSNRIWNEED 60 ELTVLKGLVD YLDKEGVQPN SDWDAFYHFL GDSIAEKFSK QQLLSKIRKL KKKFLLHMEK 120 TSQGDDMLCD SEAIRLSRMI WGQNGSESAN NQSKSFNQDE ALKDNDQVAN AEANENVAAL 180 GDKNTDVDES GTLRDAFESI MSRWLSDHQK KVQLENLMNL EAGKRKELSD EWKELCAEEA 240 RLNIKKFKYS AKFAEAAAKR * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 21 | 42 | RKRASNPSPASETTKKKKKKKK |
| 2 | 36 | 42 | KKKKKKK |
| 3 | 36 | 45 | KKKKKKKTKK |
| 4 | 37 | 45 | KKKKKKTKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | DNA-binding protein, which specifically recognizes the GL1 enhancer sequence (PubMed:12535344). May be involved in leaf initiation (PubMed:12535344). May play redundant roles with GPL1 and GPL2 in cytokinin responses by regulating the transcript levels of type-A ARR response genes (PubMed:18162594). Involved in stress responses (PubMed:21875893). Plays a repressive role in cell expansion by counteracting the positive role of CPR5 in this process, but does not regulate cell proliferation or endoreduplication (PubMed:21875893). May play a role in plant defense (PubMed:29192025). {ECO:0000269|PubMed:12535344, ECO:0000269|PubMed:21875893, ECO:0000269|PubMed:29192025, ECO:0000305|PubMed:18162594}. | |||||
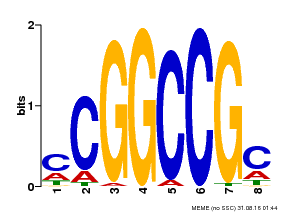
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00061 | PBM | Transfer from AT4G00270 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Thhalv10027890m |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by KNAT1. Not regulated by gibberellins. {ECO:0000269|PubMed:12535344}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006405926.1 | 0.0 | GLABROUS1 enhancer-binding protein | ||||
| Refseq | XP_006405927.1 | 0.0 | GLABROUS1 enhancer-binding protein | ||||
| Swissprot | Q9ASZ1 | 2e-71 | STKLO_ARATH; GLABROUS1 enhancer-binding protein | ||||
| TrEMBL | V4LAC3 | 0.0 | V4LAC3_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006405926.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6117 | 17 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00270.1 | 2e-67 | DNA-binding storekeeper protein-related transcriptional regulator | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Thhalv10027890m |
| Entrez Gene | 18022700 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



