 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp1g15660 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 301aa MW: 33238.6 Da PI: 8.5979 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.1 | 8.4e-13 | 5 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT+eE+e l+ +v ++G+g Wk I r +R+ ++k++w+++
Tp1g15660 5 KLKWTAEEEEALLAGVGKHGPGKWKNILRDPEfadqlSNRSNIDLKDKWRNL 56
689**********************************9************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.475 | 1 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.15E-15 | 3 | 59 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.1E-8 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.3E-14 | 5 | 56 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 3.40E-20 | 7 | 56 | No hit | No description |
| Pfam | PF13921 | 1.1E-8 | 8 | 65 | No hit | No description |
| Gene3D | G3DSA:1.10.10.10 | 1.0E-12 | 124 | 202 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00526 | 9.8E-18 | 124 | 190 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 3.59E-15 | 125 | 206 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PROSITE profile | PS51504 | 22.692 | 126 | 202 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 1.2E-11 | 128 | 200 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 301 aa Download sequence Send to blast |
MGNQKLKWTA EEEEALLAGV GKHGPGKWKN ILRDPEFADQ LSNRSNIDLK DKWRNLSVAP 60 DIQGSKDKVR TPKIKAAAFH LAAAAAAATS TPTPTSTSSP VAPLPRSGSS DLSIDDSCNI 120 MADAKNAPRY DGMIFEALST LTDANGSDVT AIFNFIEQKR HEVPPNFKRI LSSRLRRLAS 180 QGKLEKVSQL KSGTQNFYKM NDNSFLAIRT PLVSRPKETN VKPRQPNSPV PVISQEKVAH 240 ASRTAAYKLF EVDEKSEAAK AAVEEKERMI ELAERAEYIL LLAEELHEQC KEDSYSQCCS 300 I |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
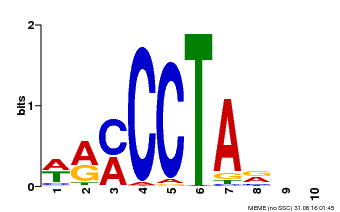
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp1g15660 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024008305.1 | 1e-162 | telomere repeat-binding factor 4 isoform X1 | ||||
| Swissprot | F4I7L1 | 1e-136 | TRB4_ARATH; Telomere repeat-binding factor 4 | ||||
| TrEMBL | V4KYH5 | 1e-155 | V4KYH5_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006416694.1 | 1e-156 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2321 | 28 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17520.1 | 1e-110 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



