 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp1g18680 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1041aa MW: 114667 Da PI: 8.4026 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.5 | 1.2e-40 | 133 | 209 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqv++C++dls+ak+yhrrhkvCevhska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk++
Tp1g18680 133 MCQVDNCTHDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTT 209
6**************************************************************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.3E-33 | 127 | 195 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.017 | 131 | 208 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.1E-37 | 132 | 210 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.5E-28 | 134 | 207 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 5.35E-7 | 792 | 930 | No hit | No description |
| SuperFamily | SSF48403 | 2.03E-5 | 838 | 937 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.0E-4 | 838 | 931 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1041 aa Download sequence Send to blast |
MDEVGAQVAA PIFIHQSLSP MGRKRNLYHQ MPNRLVPSQP QPQRRDEWDS KMWDWDSRRF 60 EAKPVDAEVL RLGSEARVDS TSRNRNGAKE GEERGLDLNL GSCVNAVEDT TQAARPSKKV 120 RSGSPGGGGN RPMCQVDNCT HDLSHAKDYH RRHKVCEVHS KATKALVGKQ MQRFCQQCSR 180 FHLLSEFDEG KRSCRRRLAG HNRRRRKTTL PDEIASGVVG PGNRDNTSNT NMDLMALLTA 240 LACAQGKNEV KPMGSSAVPE REQLLQILNK INALPLPMDL VSKLNNIGSL ARKNLDHPVV 300 NTQNDMNGAS PSTMDLLAVL SATLGSSSPD ALAILSQGGF GTKDSDKTKS SSYDRCATTN 360 LERRTVGGER SSSSNQSPSQ DSDSHAQDTR SSLSLQLFTS SPEDESRPAV ASSRKYYSSA 420 SSNPVVDRSP SSSPVMQELF PLQTSPETMR SKNHRNTSPA RPGGCLPLEL FGASNRGAAN 480 PNFKGFGQQS GYASSGSDYS PPSLNSDAQD RTGKIVFKLL DKDPSQLPGT LRTEIYNWLS 540 NIPSEMESYI RPGCVVLSVY VAMSPAAWEQ LEQNLLQRVG VMLQDSYSGF WRNARFIVNT 600 GRQLASHKNG RIRCSKSWRT WNSPELISVS PVAVVAGEET SLVVRGRSLT NDGISVRCTQ 660 TGSYVSMEVT GAACKRAIFD ELNVNSFKVK NAQPGFLGRC FIEVENGFRC DSFPLIIANA 720 SICKELNCLE EEFHPKSQDM TEELTQTSDR CPTSREEILC FLNELGWLFQ KNQTSEPQGQ 780 SDFSLSRFKF LLICSVERDY CALIRTLLDM LVERNLMNDE LSREALDMLA EIQLVSRAVQ 840 RKSTKMVELL IHYCFVTPAA GSSSKKFVFL PNITGPGGIS PLHLAACTSG SDDLVDLLTN 900 DPQEIGLSSW NSLCDATGQT PYSYAAMRNN HTYNSLVARK LADKRNKQVS LNVESEIVDH 960 MGLSRRLSSE TNKSSCATCA TAALKYRRKG SGSHRLFPTP IIHSMLAVAT VCVCVCVFMH 1020 AFPIVRQGSH FSWGGLDYGS I |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-31 | 123 | 207 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
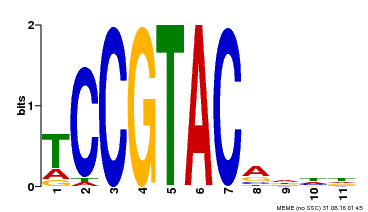
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp1g18680 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353554 | 0.0 | AK353554.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-52-I08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006416342.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | V4KAJ1 | 0.0 | V4KAJ1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006416342.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



