 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp2g20210 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 624aa MW: 71767.7 Da PI: 7.1045 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 49.9 | 8.3e-16 | 97 | 162 | 2 | 72 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrt 72
W+ +evlaL+++r+ +e+++ + We+ s+k++e gf+rsp++Ckek+e+ ++ry + +++ ++++
Tp2g20210 97 WCSDEVLALLRFRSTVENWFPEF-----TWEHTSRKLAEVGFKRSPQECKEKFEEEERRYFNSNNNTNDHH 162
********************998.....9*******************************99999888874 PP
| |||||||
| 2 | trihelix | 99.1 | 3.8e-31 | 455 | 549 | 1 | 86 |
trihelix 1 rWtkqevlaLiearr..........emeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++evlaLi++rr ++ + +++++ plWe++skkm e+g++rs+k+Ckekwen+nk+++k+k+ kkr + +s+tcpyf+ql+
Tp2g20210 455 RWPRDEVLALINIRRsissmndddlHKDGISLSSSKAVPLWERISKKMLETGYKRSAKRCKEKWENINKYFRKTKDVHKKR-PLDSRTCPYFHQLT 549
8**************5555433322112222234699*******************************************8.9***********98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 8.4E-11 | 95 | 161 | No hit | No description |
| PROSITE profile | PS50090 | 5.482 | 96 | 148 | IPR017877 | Myb-like domain |
| Pfam | PF13837 | 2.1E-19 | 454 | 550 | No hit | No description |
| CDD | cd12203 | 2.11E-25 | 454 | 529 | No hit | No description |
| PROSITE profile | PS50090 | 6.922 | 455 | 522 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 624 aa Download sequence Send to blast |
MFNGGVPEQI HRFIASPQPP PPLPPHQPAT ERSLPFPVSF ASFNSNQQAQ HMLSLDNRKI 60 IHHHHHHDIK EGGATSEWMG HTDHDSDTPR HHHHHPWCSD EVLALLRFRS TVENWFPEFT 120 WEHTSRKLAE VGFKRSPQEC KEKFEEEERR YFNSNNNTND HHMSNYNNKG NYRIYSEVEE 180 FYHHGHDDDE HVVSSEVGDN QNKRNDSLEG KGNAEETGQD LLEEGKSDHQ DQGQVEESSM 240 GNNINSNTDN VGKVGNVEDG SKSSSSSSLR DKKKRKRKKK KERFGVLKGF CEGLVRNMIA 300 QQEEMHKKLL EDMVKKEEEK MVREEAWKKQ EMDRLNKEVE IRAHEHAMAS DRNTNIIEFI 360 SKFTDNDNHD DGNYKLQSPS PSQDSSSLLL PKPQGNMKFH TSSSLLPQTL TPHNPLHSLQ 420 TNHESVEPIS TKTLKTKTQN PKPPKSDDKS DLGKRWPRDE VLALINIRRS ISSMNDDDLH 480 KDGISLSSSK AVPLWERISK KMLETGYKRS AKRCKEKWEN INKYFRKTKD VHKKRPLDSR 540 TCPYFHQLTA LYSQPSTGTT TTVTDTSAGE LETRPEENRV GSGDPDIPAV MHVDADGAGE 600 KKQFNTIYIY ITSIISFFLL FFRL |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 272 | 277 | KKRKRK |
| 2 | 272 | 279 | KKRKRKKK |
| 3 | 272 | 280 | KKRKRKKKK |
| 4 | 273 | 281 | KRKRKKKKE |
| 5 | 274 | 278 | RKRKK |
| 6 | 274 | 280 | RKRKKKK |
| 7 | 274 | 281 | RKRKKKKE |
| 8 | 276 | 281 | RKKKKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
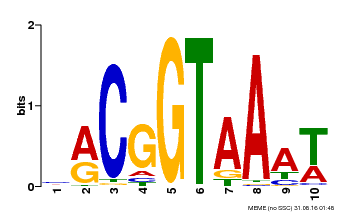
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp2g20210 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013677968.1 | 0.0 | trihelix transcription factor GTL2 | ||||
| Swissprot | Q8H181 | 0.0 | GTL2_ARATH; Trihelix transcription factor GTL2 | ||||
| TrEMBL | A0A078IRA5 | 0.0 | A0A078IRA5_BRANA; BnaC07g49430D protein | ||||
| STRING | Bo2g152980.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8268 | 28 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 0.0 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



