 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp3g11190 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 444aa MW: 49379 Da PI: 6.2037 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.8 | 8.8e-32 | 237 | 291 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr+rWtpeLHe+F++ v++L G+ekAtPk++l+lm+v+gLt++hvkSHLQkYRl
Tp3g11190 237 KPRMRWTPELHESFLNSVNKLEGPEKATPKAVLKLMNVEGLTIYHVKSHLQKYRL 291
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-29 | 235 | 292 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.2E-16 | 236 | 291 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.9E-24 | 237 | 292 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.5E-8 | 239 | 290 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.5E-22 | 327 | 374 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 444 aa Download sequence Send to blast |
MNRHRLVSTA HDECNKGIRQ ACSSSLSPAH NFLNVQPETM KSPFIRSQSP DSPGQRWPKN 60 SSQGTFSRSS TFCTNLYLSS SSTSETQKHM GNSLPFLPDP SAYSHSASGV ESARSPYVFS 120 EDLGNPFDGD NSGSLVKDFL NLSGDGCSNG GFHDLDCSND SYCLSDQMEL QFLSDELELA 180 ITDRAETPRL DEIYETPLAS SNQVTGLSPS QRCVAGAMSI DVVSSHPSPG SAAANHKPRM 240 RWTPELHESF LNSVNKLEGP EKATPKAVLK LMNVEGLTIY HVKSHLQKYR LAKYMPEKKE 300 EKKNVNSEVK RLALSNSEVD EKKRGAIQLT EALRMQMEVQ KQLHEQLEVQ RVLQLRIEEH 360 AKYLEKMLEE QRKTGRLISS SSSQTLISPS DDSIPDCQNM SKAEVSMPQA SSSEKNIASE 420 TEDDKCESPQ KRRRLENKAE SEIS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-24 | 237 | 294 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 430 | 434 | KRRRL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
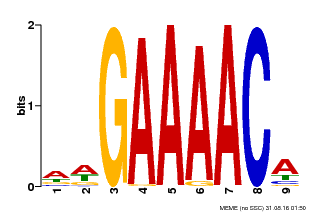
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp3g11190 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY050889 | 0.0 | AY050889.1 Arabidopsis thaliana unknown protein (At3g13040) mRNA, complete cds. | |||
| GenBank | AY096674 | 0.0 | AY096674.1 Arabidopsis thaliana unknown protein (At3g13040) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020886314.1 | 0.0 | myb family transcription factor PHL6 | ||||
| Swissprot | Q949U2 | 0.0 | PHL6_ARATH; Myb family transcription factor PHL6 | ||||
| TrEMBL | D7L0M9 | 0.0 | D7L0M9_ARALL; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.3__1373__AT3G13040.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6786 | 27 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 0.0 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



