 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp3g31590 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 168aa MW: 18555.5 Da PI: 4.7404 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 40.1 | 7.7e-13 | 28 | 86 | 4 | 62 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
+++ +r+ +NRe+ArrsR RK++ ++ L+ + +L++ N++ l ++++ k ++e
Tp3g31590 28 DRKRKRMLSNRESARRSRMRKQKHVDDLTAQINQLSSDNRQILTSLTVTSQLYMKIQAE 86
7899***********************************99887777777777666665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.9E-16 | 25 | 89 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.6E-10 | 27 | 72 | No hit | No description |
| PROSITE profile | PS50217 | 10.059 | 27 | 90 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.1E-9 | 28 | 79 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.12E-12 | 29 | 80 | No hit | No description |
| CDD | cd14702 | 7.23E-22 | 30 | 81 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 32 | 47 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 168 aa Download sequence Send to blast |
MASSSSTYRS SSSSDGGNPS DSVVAVDDRK RKRMLSNRES ARRSRMRKQK HVDDLTAQIN 60 QLSSDNRQIL TSLTVTSQLY MKIQAENSIL TAQMSELSTR LESLNEIVDL VTTTNGAGFD 120 QIDGCGFDDR TVGINSDGYY DDMMSGVNPW GGSVYTNQPI MANDINMY |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 28 | 49 | RKRKRMLSNRESARRSRMRKQK |
| 2 | 41 | 48 | RRSRMRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to specific DNA sequences in target gene promoters. BZIP2-BZIP63-KIN10 complex binds to the ETFQO promoter to up-regulate its transcription (PubMed:29348240). {ECO:0000250|UniProtKB:O65683, ECO:0000269|PubMed:29348240}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00267 | DAP | Transfer from AT2G18160 | Download |
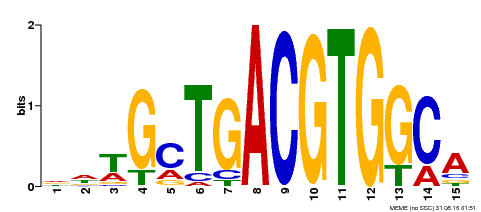
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp3g31590 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353485 | 0.0 | AK353485.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-45-P17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006409191.1 | 1e-102 | bZIP transcription factor 2 | ||||
| Swissprot | Q9SI15 | 1e-92 | BZIP2_ARATH; bZIP transcription factor 2 | ||||
| TrEMBL | A0A3P6ES75 | 1e-105 | A0A3P6ES75_BRAOL; Uncharacterized protein | ||||
| STRING | Bo7g011520.1 | 1e-105 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM501 | 28 | 154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18160.1 | 3e-89 | basic leucine-zipper 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



