 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp4g08390 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 429aa MW: 48464.5 Da PI: 4.8921 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.9 | 1e-17 | 22 | 69 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Edell+ +v ++G g+W+++ ++ g+ R++k+c++rw ++l
Tp4g08390 22 KGPWTLAEDELLIAYVDKHGDGNWNAVQKHSGLSRCGKSCRLRWVNHL 69
79*******************************************996 PP
| |||||||
| 2 | Myb_DNA-binding | 45.3 | 2.1e-14 | 75 | 118 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++T+ E+++ +++++ +G++ W++ a++++ gRt++++k++w++
Tp4g08390 75 KGSFTEKEEQRVIELHASMGNK-WARMAAELP-GRTDNEIKNFWNT 118
799*******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.542 | 17 | 69 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.0E-15 | 21 | 71 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.41E-28 | 21 | 116 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.6E-16 | 22 | 69 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-23 | 23 | 76 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.27E-12 | 24 | 69 | No hit | No description |
| PROSITE profile | PS51294 | 24.476 | 70 | 124 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-14 | 74 | 122 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-13 | 75 | 118 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-24 | 77 | 122 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.97E-10 | 77 | 120 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MRKVRHDSGS GEDSALEKTF QKGPWTLAED ELLIAYVDKH GDGNWNAVQK HSGLSRCGKS 60 CRLRWVNHLR PDLKKGSFTE KEEQRVIELH ASMGNKWARM AAELPGRTDN EIKNFWNTRL 120 KRLQRLGLPI YPDEVQEQAM NAAAQSGQNT DSLNGHHSQE SLELDCLEIP EVEFKHLQLN 180 NCTSLYHSML RNFPKGNFMR QRPCLYQPNT YSVIGSPYMS PNRKRFRESE TPFPYTGGYA 240 TNEQSAQLWN NPFVESTSEQ FNVLPDSHFL GNATYSSSSE ALIHEAEKLE LPSFQCFDPQ 300 EEPGDWETQL SNPMPALESD NTLVQSPLTD QTPPNFPSSF YDGLLESVVY GSSGERRETN 360 TDPDSPLLQS SLLGHIELTP ATADTTGHNS NFETDLGQFG PISSNESRTS FDGGDWIRYL 420 LGEDRAYTK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 4e-29 | 22 | 122 | 7 | 106 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00279 | DAP | Transfer from AT2G26960 | Download |
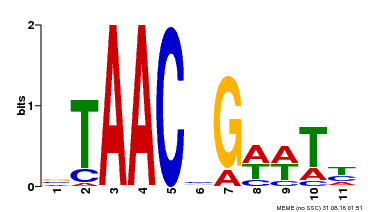
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp4g08390 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006408602.2 | 0.0 | transcription factor MYB104 | ||||
| Swissprot | Q9SM27 | 1e-124 | MY104_ARATH; Transcription factor MYB104 | ||||
| TrEMBL | A0A078HJC0 | 0.0 | A0A078HJC0_BRANA; BnaC03g26620D protein | ||||
| TrEMBL | A0A0D3B7F2 | 0.0 | A0A0D3B7F2_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6AYS0 | 0.0 | A0A3P6AYS0_BRAOL; Uncharacterized protein | ||||
| STRING | Bo3g041750.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9198 | 21 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G26960.1 | 1e-179 | myb domain protein 81 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



