 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp4g12830 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 398aa MW: 44308.9 Da PI: 7.274 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 95 | 5.2e-30 | 169 | 226 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+Dgy WrKYGqK+vk+se+prsY++Ct+++C +kk ve+++ d++++ei+Y+g Hnh+k
Tp4g12830 169 NDGYGWRKYGQKQVKKSENPRSYFKCTYPNCVSKKIVETAS-DGQITEIIYKGGHNHPK 226
8***************************************9.***************85 PP
| |||||||
| 2 | WRKY | 103.2 | 1.4e-32 | 334 | 390 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+vkg+++prsYY+Ct++gC v+k+versaed ++v +tYeg+Hnh+
Tp4g12830 334 DGFRWRKYGQKVVKGNTNPRSYYKCTHQGCGVRKQVERSAEDDRAVLTTYEGRHNHD 390
9*******************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.9E-25 | 157 | 227 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.71E-23 | 165 | 227 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.3E-30 | 168 | 226 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-22 | 169 | 225 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 20.461 | 169 | 227 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.6E-34 | 317 | 392 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.31E-28 | 324 | 392 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 34.004 | 327 | 392 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.0E-38 | 332 | 391 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.5E-25 | 334 | 390 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 398 aa Download sequence Send to blast |
MSSTSFTDLL ASSGVDCYEE DEDFRVSGSV SLYQNGGFCP EKTGSGLPKF KTAQPPPLPI 60 SQSPHSFAFS DLLDSPLLLS SSHSLISPTT GAFPYQGLNG TNNHSGFPWQ LQSQSQPQPL 120 NASSALQETC GAQDLQKKQD PVPREFAAHS FGSDRQIKVP SYMVSRNSND GYGWRKYGQK 180 QVKKSENPRS YFKCTYPNCV SKKIVETASD GQITEIIYKG GHNHPKPEFT KRPSGSTSIP 240 SSSANARRMF NPSSVVSETH DQSENSSISF DYSDLEQKSF KSEYGEIDEE NEQPETKRLK 300 REGEDEGMSV EVSRGVKEPR VVVQTISEID VLIDGFRWRK YGQKVVKGNT NPRSYYKCTH 360 QGCGVRKQVE RSAEDDRAVL TTYEGRHNHD IPTALRRS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-32 | 323 | 392 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-32 | 323 | 392 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Functions with WRKY33 as positive regulator of salt stress response and abscisic acid (ABA) signaling (PubMed:18839316). Plays a partial role in heat stress tolerance (PubMed:19125253). Functions with WRKY26 and WRKY33 as positive regulator of plant thermotolerance by partially participating in ethylene-response signal transduction pathway (PubMed:21336597). {ECO:0000250|UniProtKB:Q9SI37, ECO:0000269|PubMed:18839316, ECO:0000269|PubMed:19125253, ECO:0000269|PubMed:21336597}. | |||||
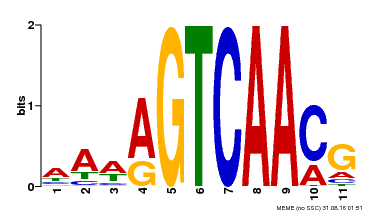
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00282 | DAP | Transfer from AT2G30250 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp4g12830 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt stress (PubMed:18839316). Induced by heat stress (PubMed:19125253). {ECO:0000269|PubMed:18839316, ECO:0000269|PubMed:19125253}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK352934 | 0.0 | AK352934.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-12-D15. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006410139.1 | 0.0 | probable WRKY transcription factor 25 | ||||
| Swissprot | O22921 | 0.0 | WRK25_ARATH; Probable WRKY transcription factor 25 | ||||
| TrEMBL | E4MWN7 | 0.0 | E4MWN7_EUTHA; mRNA, clone: RTFL01-12-D15 | ||||
| TrEMBL | V4M953 | 0.0 | V4M953_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006410139.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM11603 | 17 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G30250.1 | 0.0 | WRKY DNA-binding protein 25 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



