 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp4g20580 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 344aa MW: 39404.9 Da PI: 8.9564 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.3 | 6.3e-32 | 55 | 108 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+ave+LGG+e+AtPk ++++m++kgL+++hvkSHLQ+YR+
Tp4g20580 55 PRLRWTPDLHLRFVRAVERLGGQERATPKLVRQMMNIKGLSIAHVKSHLQMYRS 108
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.273 | 51 | 111 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.6E-29 | 51 | 109 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.97E-14 | 53 | 109 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-22 | 55 | 110 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.2E-9 | 56 | 107 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MISSMEGGEK ADREEDEEEE EEEGEREESK VSSNSTVEEI DKKTKVRPYV RSKVPRLRWT 60 PDLHLRFVRA VERLGGQERA TPKLVRQMMN IKGLSIAHVK SHLQMYRSKK MDDQGQVIAD 120 HRHFMETSTD RNVYKLSQLP MFRGYNINHN HDSPFRYGSK LSNVSLWNSS SHETNRSLID 180 SPGLIRGSSI SNNIRGNDYS TNNRSFQNIV YSSSISNHLP KLRHNHQERA NSVTFNSIQE 240 HSRTFEKFKI GLEESTNHAY CNKTRGKRNA STSIDLDLDL SLNLRVPEKT IMEETETAAT 300 TTDQRLSLSL CSGSSSWKKS RVIKTDEEDR TVKIGQASTL DLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 9e-16 | 56 | 110 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-16 | 56 | 110 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-16 | 56 | 110 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-16 | 56 | 110 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
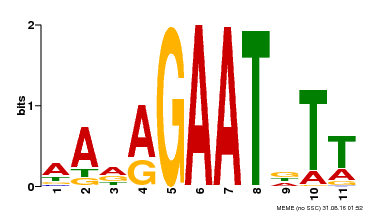
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp4g20580 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_181364.2 | 0.0 | myb-like HTH transcriptional regulator family protein | ||||
| TrEMBL | A0A178VYP6 | 0.0 | A0A178VYP6_ARATH; Uncharacterized protein | ||||
| TrEMBL | Q8GYE4 | 0.0 | Q8GYE4_ARATH; Myb-like HTH transcriptional regulator family protein | ||||
| STRING | AT2G38300.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM363 | 28 | 184 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 1e-168 | G2-like family protein | ||||



