 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp4g27690 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 222aa MW: 25472.6 Da PI: 8.7155 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 23.3 | 8.3e-08 | 9 | 26 | 1 | 18 |
S---SHHHHHHHHHHHHH CS
SRF-TF 1 krienksnrqvtfskRrn 18
krienk+nrqvtfskRrn
Tp4g27690 9 KRIENKINRQVTFSKRRN 26
79***************8 PP
| |||||||
| 2 | K-box | 77.4 | 3.7e-26 | 61 | 139 | 5 | 83 |
K-box 5 sgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkk 83
+++ e+ +++++qe++kLk+++e+L r++RhllGedL+++++keLq Le+qLe +l+ R++K++ l + ++l+ k
Tp4g27690 61 LSNNRPEESTQNWCQEVTKLKSKYESLVRTNRHLLGEDLGEMNVKELQALERQLEAALTATRQRKERQLGDINKQLKIK 139
455578888************************************************************9999999877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 15.6 | 1 | 45 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.5E-16 | 1 | 44 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 1.7E-18 | 2 | 73 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 6.9E-5 | 10 | 26 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.7E-21 | 67 | 139 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 11.7 | 70 | 168 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048437 | Biological Process | floral organ development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 222 aa Download sequence Send to blast |
MGRGRVEMKR IENKINRQVT FSKRRNVALI VFSSRGKLYE FGSVGVEKTI ERYHRCYNCS 60 LSNNRPEEST QNWCQEVTKL KSKYESLVRT NRHLLGEDLG EMNVKELQAL ERQLEAALTA 120 TRQRKERQLG DINKQLKIKF ETEGHAFKSF QDLWTNSAAS VVGDAHNSEF PVQSSHPDSM 180 DCNTEPFLQI GFQQHYYVQG EGSSVSKSNA ACETNFVQGW VL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development. | |||||
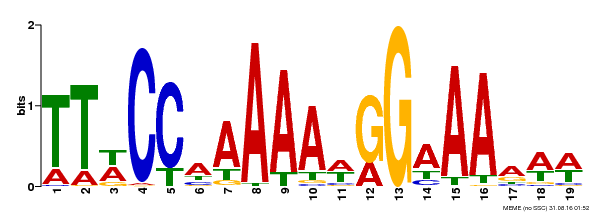
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp4g27690 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ508055 | 1e-124 | AJ508055.1 Brassica oleracea var. botrytis mRNA for MADS-box protein AGL6-a (agl6-a gene), long splice variant. | |||
| GenBank | AJ508409 | 1e-124 | AJ508409.1 Brassica oleracea var. botrytis mRNA for MADS-box protein AGL6-a (agl6-a gene), short splice variant. | |||
| GenBank | KC984301 | 1e-124 | KC984301.2 Brassica oleracea var. viridis AGL6 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_182089.1 | 1e-144 | AGAMOUS-like 6 | ||||
| Swissprot | P29386 | 1e-145 | AGL6_ARATH; Agamous-like MADS-box protein AGL6 | ||||
| TrEMBL | Q1PEU3 | 1e-142 | Q1PEU3_ARATH; AGAMOUS-like protein 6 | ||||
| STRING | AT2G45650.1 | 1e-143 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4302 | 26 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 1e-133 | AGAMOUS-like 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



