 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp4g29060 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassicaceae incertae sedis; Schrenkiella
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 929aa MW: 104015 Da PI: 6.1374 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135.6 | 1.6e-42 | 143 | 220 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+Ceadls++k+yhrrhkvCe+hska++++v g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Tp4g29060 143 VCQVENCEADLSKVKDYHRRHKVCEMHSKATSAIVGGIMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 220
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.3E-34 | 137 | 205 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.616 | 141 | 218 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.62E-39 | 143 | 222 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.3E-31 | 144 | 217 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 1.71E-7 | 717 | 818 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.0E-7 | 718 | 819 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.41E-9 | 720 | 816 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 929 aa Download sequence Send to blast |
MEARIEGEPQ HFYGFRAMDL RSVGKKSVEW DLNDWKWDGD LFVATQLNPG GSETMGRQLF 60 PLGNSSNSSS SCSDEGNNTE LTVTRKREIE KKRRAAATGE HNNGGGLTLK LGDNGYDLNG 120 EREGNSAAKK TKFGGGGVTN RSVCQVENCE ADLSKVKDYH RRHKVCEMHS KATSAIVGGI 180 MQRFCQQCSR FHVLQEFDEG KRSCRRRLAG HNKRRRKTNP EPGTNGNPTS DDHQSSNYLL 240 ISLLKILSNM HYNPGDQDLM SHLLKSLVSH AGEQLGKNLV ELLLQGRSQA SLNIGNSGLL 300 AIEQATQEDL KQAPEIPRQE LYANGTATEN RLEKQVKVND FDLNDIYIDS DDGTDIERSP 360 PPTNPATSSL DYPSWVHQSS PPQTSRNSDS ASDQSPSSSS EDAQMRTGRI VFKLFGKEPN 420 DFPVVLRGQI LDWLSHSPTD MESYIRPGCI VLTIYLRQAE TAWEELSDDL GFSLRKLLDL 480 SDDPLWTTGW IYVRVQNQLA FLYNGQVAID TSLPLRSRDY GHIISVRPLA VAATRKAQFT 540 VKGINLRRPG TSLLCAVEGK YLIQETKHDS TKENDDLKEN NEIVESVTFS CDLPITSGRG 600 FMEIEDQGLS SSFFPFIVVE DDDVCSELRI LEATLEFTGT DSAKQAMEFI HEIGWLLHRS 660 KLGESDPNPD VFPLTRFKWL IEFSMDLEWC AVIRKLLNMF FEGAVGDSSS DSELSELCLL 720 HRAVRKNSKP MVEMLLRYAP KQQRHSLFRP DDAGPAGLTP LHIAAGKDGS EDVLDALTED 780 PTMVGIEAWR TSRDSTGFTP EDYARLRGHF SYIHLIQRKI NKKSTTEGHV VVNIPVSFSE 840 REQKEESKSG LMASALEVTH TNHQLQCKLC DHKLVYGTAR RSVAYRPAML SMVAIAAVCV 900 CVALLFKSCP EVLYVFQPFR WELLDYGTR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 8e-33 | 134 | 217 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
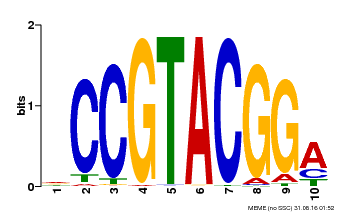
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp4g29060 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC815948 | 0.0 | KC815948.1 Arabis alpina SQUAMOSA promoter binding protein-like protein 1 (SPL1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006397898.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | V4KN69 | 0.0 | V4KN69_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006397898.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



