 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA18393 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 269aa MW: 30716.1 Da PI: 8.9522 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.3 | 1.5e-31 | 34 | 84 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krienk+nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fs++gklye++s
Tp57577_TGAC_v2_mRNA18393 34 KRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSNRGKLYEFCS 84
79***********************************************96 PP
| |||||||
| 2 | K-box | 97.5 | 2e-32 | 109 | 199 | 9 | 99 |
K-box 9 leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenk 92
+ea + s+qqe+ kLk+++e+Lqr+qR+l+GedL++Ls k+L+ Le+qL++slk+iRs++++++l+q+ +lq+ke+ l e+n+
Tp57577_TGAC_v2_mRNA18393 109 SKEALELSSQQEYLKLKARYESLQRSQRNLMGEDLGPLSSKDLETLERQLDSSLKQIRSTRTQFMLDQLGDLQRKEHLLCEANR 192
455666789*************************************************************************** PP
K-box 93 aLrkkle 99
aLr+++e
Tp57577_TGAC_v2_mRNA18393 193 ALRQRME 199
***9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.1E-41 | 26 | 85 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.487 | 26 | 86 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 1.57E-33 | 27 | 102 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.95E-44 | 27 | 101 | No hit | No description |
| PRINTS | PR00404 | 1.1E-32 | 28 | 48 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 28 | 82 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.8E-26 | 35 | 82 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.1E-32 | 48 | 63 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.1E-32 | 63 | 84 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 15.059 | 114 | 204 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 3.9E-26 | 116 | 198 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MYDLPTTRRV HLLKRIGEQV LCSEKMGRGR VELKRIENKI NRQVTFAKRR NGLLKKAYEL 60 SVLCDAEVAL IVFSNRGKLY EFCSTSSMLK TLERYQKCNY GAPEANVSSK EALELSSQQE 120 YLKLKARYES LQRSQRNLMG EDLGPLSSKD LETLERQLDS SLKQIRSTRT QFMLDQLGDL 180 QRKEHLLCEA NRALRQRMEG YQINSLQLNL SAEDMGYGRH HHQVHNQGDD LFQPIECQPT 240 LQIGYQGDPG SVVTAGPSMN NFMGGWLP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 6e-45 | 100 | 198 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 6e-45 | 100 | 198 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 6e-45 | 100 | 198 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 6e-45 | 100 | 198 | 1 | 101 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000269|PubMed:9869431}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00605 | ChIP-seq | Transfer from AT1G24260 | Download |
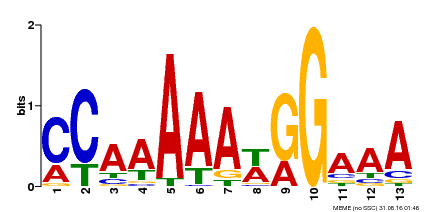
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA18393 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ223318 | 0.0 | AJ223318.1 Pisum sativum mRNA for MADS-box transcription factor. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003601749.1 | 1e-166 | MADS-box transcription factor 1 isoform X2 | ||||
| Swissprot | O65874 | 1e-167 | MTF1_PEA; MADS-box transcription factor 1 | ||||
| TrEMBL | G7JBE5 | 1e-165 | G7JBE5_MEDTR; MADS-box transcription factor | ||||
| STRING | AES72000 | 1e-166 | (Medicago truncatula) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G24260.1 | 1e-126 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA18393 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



