 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Tp57577_TGAC_v2_mRNA32765 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Trifolium
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 300aa MW: 33069 Da PI: 10.4089 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 345.4 | 1.3e-105 | 13 | 299 | 1 | 301 |
GAGA_bind 1 mdddgsre.rnkgyyepaaslkenlglqlmssiaerdakirernlalsekkaavaerd.........maflqrdkalaernkal 74
md d+ + n+gyyep +s+k++lglqlm s++e+ ++ + ls++++++ +rd m++ +rd+++++
Tp57577_TGAC_v2_mRNA32765 13 MDGDNGLNiSNWGYYEPVSSFKSHLGLQLMPSMPEKPLLGSHNAAVLSGSNGPFRNRDiglpqttypMDY-MRDAWISS----- 90
8888888889************************998766555555************************.********..... PP
GAGA_bind 75 verdnkllalllvenslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakk 158
+rdnk++++ ++ ++ p+g++++++++s++ +q+++ p+l ++ e+ p+eea+ ++k + + kkrq +k+
Tp57577_TGAC_v2_mRNA32765 91 -QRDNKYMNMNMIPTN-----PPGYSSIPEASSAHPIQMIRPPEL------VK-----EERPTEEAPVVEKSNGTGKKRQGPKV 157
.9********999886.....*****************9332222......22.....345666667777888888899999** PP
GAGA_bind 159 pkekkakkkkkksekskkkvkkesaderskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvy 242
pk++kakk+k+ ++++k +++++s++++ +a+kk++++++ng++lD+s++P+PvCsCtG+++qCY+WG+GGWqSaCCtt+iS+y
Tp57577_TGAC_v2_mRNA32765 158 PKSPKAKKPKRGPRVPK-DENSPSVQRTPRAPKKTTEIAINGIDLDISSIPIPVCSCTGTPQQCYRWGSGGWQSACCTTAISIY 240
***********888875.55666667899******************************************************* PP
GAGA_bind 243 PLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
PLP+stkrrgaRiagrKmS gafkk+LekLaaeGy++snp+DL+++WAkHGtnkfvtir
Tp57577_TGAC_v2_mRNA32765 241 PLPMSTKRRGARIAGRKMSIGAFKKVLEKLAAEGYNFSNPIDLRTYWAKHGTNKFVTIR 299
**********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 6.0E-105 | 13 | 299 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 5.1E-156 | 13 | 299 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 300 aa Download sequence Send to blast |
LKQKFSSTFS VVMDGDNGLN ISNWGYYEPV SSFKSHLGLQ LMPSMPEKPL LGSHNAAVLS 60 GSNGPFRNRD IGLPQTTYPM DYMRDAWISS QRDNKYMNMN MIPTNPPGYS SIPEASSAHP 120 IQMIRPPELV KEERPTEEAP VVEKSNGTGK KRQGPKVPKS PKAKKPKRGP RVPKDENSPS 180 VQRTPRAPKK TTEIAINGID LDISSIPIPV CSCTGTPQQC YRWGSGGWQS ACCTTAISIY 240 PLPMSTKRRG ARIAGRKMSI GAFKKVLEKL AAEGYNFSNP IDLRTYWAKH GTNKFVTIR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
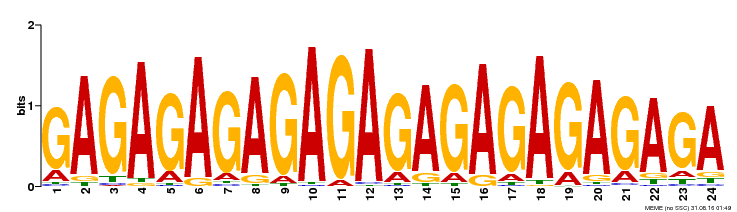
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Tp57577_TGAC_v2_mRNA32765 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT142719 | 0.0 | BT142719.1 Medicago truncatula clone JCVI-FLMt-15C3 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004513327.1 | 0.0 | protein BASIC PENTACYSTEINE1-like | ||||
| Refseq | XP_004513328.1 | 0.0 | protein BASIC PENTACYSTEINE1-like | ||||
| Swissprot | Q9SKD0 | 5e-91 | BPC1_ARATH; Protein BASIC PENTACYSTEINE1 | ||||
| TrEMBL | A0A2K3N7D6 | 0.0 | A0A2K3N7D6_TRIPR; Protein basic pentacysteine1-like (Fragment) | ||||
| STRING | XP_004513327.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2370 | 32 | 83 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01930.2 | 3e-86 | basic pentacysteine1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Tp57577_TGAC_v2_mRNA32765 |



